SUPPLEMENTAL MATERIAL METABOLIC HOMEOSTASIS IS MAINTAINED IN MYOCARDIAL HIBERNATION BY ADAPTIVE CHANGES IN THE TRANSCRIPTOME AND PROTEOME
|
|
|
- Τῑτάν Γιάγκος
- 5 χρόνια πριν
- Προβολές:
Transcript
1 SUPPLEMENTAL MATERIAL METABOLIC HOMEOSTASIS IS MAINTAINED IN MYOCARDIAL HIBERNATION BY ADAPTIVE CHANGES IN THE TRANSCRIPTOME AND PROTEOME Manuel Mayr 1, Dalit May 2, Oren Gordon 2, Basetti Madhu 3, Dan Gilon 4, Xiaoke Yin 1, Qiuru Xing 1, Ignat Drozdov 5, Chrysanthi Ainali 5, Sophia Tsoka 5, Qingbo Xu 1, John Griffiths 3, Anton Horrevoets 6, Eli Keshet 2 1 King s British Heart Foundation Centre, King s College London, London, UK 2 Molecular Biology, The Hebrew University-Hadassah University Hospital, Jerusalem, Israel 3 Cancer Research UK, Cambridge Research Institute, Cambridge, UK 4 Cardiology Department, Hadassah-Hebrew University Medical Center 5 Centre for Bioinformatics - School of Physical Sciences and Engineering, King s College London, London, UK 6 Molecular Cell Biology and Immunology, VU University Medical Center, Amsterdam, The Netherlands Correspondence to: Dr. Manuel Mayr, Cardiovascular Division, King's College London 125 Coldharbour Lane, London SE5 9NU, UK Phone: +44 (0) Fax: +44 (0) manuel.mayr@kcl.ac.uk
2 SUPPLEMENTAL MATERIAL AND METHODS Hypoxyprobe TM staining. Hypoxyprobe TM (pimonidazole hydrochloride [1]) staining was performed according to the manufacturer's instruction (60 mg/kg body, Chemicon). Hypoxic cells activate pimonidazole hydrochloride and bind 2- nitroimidazole compounds. Oxygen competes for the addition of the first electron to Hypoxyprobe TM, and it is believed that this competition accounts for the oxygen dependence of 2-nitroimidazole reductive activation and binding. The second part of the assay includes a primary antibody reagent that recognizes Hypoxyprobe TM adducts in hypoxic cells. To minimise non-specific background staining, a primary fluorescein (FITC)-conjugated mouse monoclonal antibody directed against pimonidazole protein adducts was used in combination with a secondary mouse anti- FITC Mab conjugated to horseradish peroxidase (HRP) as previously reported [2]. Real-time PCR (qpcr). For qpcr analysis, cdna was generated from 5 mg of total RNA by using Verso cdna kit (Thermo scientific) and the respective primers. Samples were normalized according to L19 mrna levels by SYBR green qpcr. The primer sequences were as follows: for Kir6.2 F- TCTCCATCGAGGTCCAGGTG, R CCCGACGATATTCTGCACAAT; for SUR2A F-TCTGCATCGGGTTCACACC, R-CCTGGGCCAAGAGGCTTT; for GLUT1 F- CTTTGGTCTCTCTCCGTGGC, R- AGCATGGAGTTCCGCCTG; for FOXO1 F- AATGATGGGCCCTAATTCGG, R- GCTGTGCATGTCCAGGGTG. Difference in-gel electrophoresis (DIGE). Protein extracts were prepared from homogenized hearts using a standard lysis bufffer (9M urea, 1% DTT, 4% CHAPS, 0.8% Pharmalytes 3-10, protease and phosphatase inhibitors (Complete Mini, Roche). After centrifugation at 13,000 g for 10 min, the supernatant containing soluble proteins was harvested and the protein concentration was determined using a 2
3 modification of the method described by Bradford. Solubilised samples were divided into aliquots and stored at 80 C. For DIGE, proteins were precipitated (ReadyPrep 2-D Clean-up kit, Biorad) and resuspended in DIGE buffer (30mM TrisCl ph 8.5, 8M urea, 4% w/v CHAPS). The fluorescence dye labelling reaction was carried out at a dye/protein ratio of 400 pmol/100µg. After incubation on ice for 30 min, the labelling reaction was stopped by scavenging non-bound dyes with 10mM lysine for 15 min. For two-dimensional gel electrophoresis (2-DE), extracts were loaded on nonlinear immobilized ph gradient 18-cm strips, 3-10 (GE healthcare). In addition to using a wide-range ph gradient, we separated cardiac proteins on a ph 4 to 7 gradient to increase spatial resolution for phosphoprotein staining. 100 µg per sample was applied to the IPG strip using an in-gel rehydration method. Samples were diluted in rehydration solution (8 M urea, 0.5% w/v CHAPS, 0.2% w/v DTT, and 0.2 % w/v Pharmalyte ph 3-10) and rehydrated overnight in a reswelling tray. Strips were focused at 0.05 ma/ipg strip for 35 kvh at 20 C (IPGphor, GE healthcare). Once IEF was completed the strips were equilibrated in 6M urea containing 30% v/v glycerol, 2% w/v SDS and 0.01% w/v Bromphenol blue, with addition of 1% w/v DTT for 15 min, followed by the same buffer without DTT, but with the addition of 4.8% w/v iodoacetamide for 15 min. SDS-PAGE was performed using 12% T (total acrylamide concentration), 2.6% C (degree of cross-linking) polyacrylamide gels without a stacking gel, using the Ettan DALT system (GE healthcare). The second dimension was terminated when the Bromphenol dye front had migrated off the lower end to the gels. After electrophoresis, fluorescence images were acquired using the Typhoon variable mode imager 9400 (GE healthcare). A pre-scan was performed to optimize the scanning parameters to exclude spot saturation. Differences in protein expression were analyzed using the Decyder software (version 6.5, GE healthcare). 3
4 Corrections for multiple testing were performed by the Benjamini-Hochberg equation, yielding False Discovery rates (FDR)[3]. Finally, gels were fixed overnight in methanol: acetic acid: water solution (4:1:5 v/v/v). Protein profiles were visualised by silver staining using the Plus one silver staining kit (GE healthcare) with slight modifications to ensure compatibility with subsequent mass spectrometry analysis [4]. For documentation, silver-stained gels were scanned in transmission scan mode using a calibrated scanner (GS-800, Biorad). Principal Component Analysis (PCA). For proteomics, DIGE gels were analysed using the EDA module of the Decyder software (version 6.5, GE healthcare). Principal component analysis (PCA) was performed on all differentially expressed proteins after removal of missing values to give an initial overview of the groupings. PCA replaces a group of variables with a smaller number of new variables, called principal components (PC), which are linear combinations of the original variables. Projecting the observation on one of these axes generates a new variable designed to maximize the description of the variance in the data set. For pattern analysis, hierarchical clustering was performed on the Student s T-test<0.05 dataset. LC-MS/MS. Gel pieces containing selected protein spots were treated overnight with modified trypsin (Promega) according to a published protocol modified for use with an Investigator ProGest (Genomic Solutions, Huntington, UK) robotic digestion system [5]. Following enzymatic degradation, peptides were separated by a Surveyor HPLC system on a reverse-phase column and applied online to a LCQ deca XP ion-trap mass spectrometer. Spectra were collected from the iontrap mass analyzer using full ion scan mode over the mass-to-charge (m/z) range MS-MS scans were performed on each ion using dynamic exclusion. Spectra were searched against a mouse database (UniProt 15.13, entries) using the 4
5 SEQUEST algorithm incorporated in the Bioworks Browser SP1 (Thermo Fisher Scientific). The following criteria were used: 1.5 AMU for the precusor ion, 1.0 AMU for fragment ions, one missed cleavage per peptide. Carbamidomethylation of cysteine as well as partial oxidation of methionine were assumed. Search results were imported into Scaffold (v , Proteome Software). The following filter were applied: 99.0% protein probability, 95% peptide probability, a minimum of 2 peptides. Immunoblotting. 50 µg of protein extracts were separated on 4-20% gradient gels (Novex, Invitrogen) and transferred to nitrocellulose membranes. Membranes were blocked in 5% PBS milk and probed with the following antibodies: lactate dehydrogenase (ab7639-1, 1:100, Abcam), glucose transporter 1 (GLUT-1, GT11-A, Alpha Diagnostics Intl.), IGFBP-2 (AF797, 0.1 µg/ml, R&D), peroxiredoxin 1 (LF- PA0001, 1:2000, Lab Frontier), peroxiredoxin 2 (LF-PA0007, 1:2000, Lab Frontier), SOD-1 (sc-11407, 1:100, Santa Cruz), and SOD-2 (06-984, 1:500, Upstate). After incubation with HRP-conjugated secondary antibodies and washing 3 times for 5 minutes each in 0.5% PBS tween, bands were detected on X-ray films using enhanced chemiluminescence (ECL, GE healthcare). Phosphate-affinity gel electrophoresis (Phos-tag TM ). For phosphate-affinity gel electrophoresis, 50 µg of protein extracts were separated on 10% polyacrylamide gels containing 50 μm Phos-tag TM (Wako Chemicals GmbH) and 50 μm MnCl 2 (Sigma). Gels were soaked in transfer buffer with 1mM EDTA for 10 min, then in transfer buffer for another 10 min prior to blotting onto a polyvinylidene fluoride (PVDF) membrane. Membranes were probed with antibodies to myosin light chain 2 (Abcam 89594) and cardiac troponin I (Cell Signaling Technology #4002). Proton nuclear magnetic resonance spectroscopy ( 1 H-NMR). Frozen heart 5
6 tissues were ground under liquid nitrogen using a mortar and pestle (n=5 and n=3 for for hibernating and control hearts, respectively). Water-soluble metabolites were extracted in 6% perchloric acid. Deproteinised extracts were transferred to ice cold centrifuge tubes and centrifuged at 3000 rpm for 10 minutes at 4 C. The supernatant was transferred to fresh cold centrifuge tubes and neutralised to ph 7 with 10M KOH. After neutralization, extracts were centrifuged, the supernatant was collected, freezedried and reconstituted in deuterium oxide (D 2 O). Immediately before the NMR analysis[6], the ph was readjusted to 7 with PCA or KOH. 0.5ml of the extracts were placed in 5mm NMR tubes. 1 H-NMR was performed using a Bruker 600MHz spectrometer. The water resonance was suppressed by using gated irradiation centred on the water frequency. Sodium 3-trimethylsilyl-2,2,3,3-tetradeuteropropionate (TSP) was added to the samples for chemical shift calibration and quantification. Network construction and pathway analysis. The previously published raw transcriptomics data [7] were first reanalyzed to correct for multiple testing[8]. This yielded a total of 166 induced and 195 repressed genes during hibernation at Baysian p<0.01 and a FDR<20%. The original 8.cel files were uploaded into the Bioconductor R-package ( 2 chips that did not pass QC were discarded, and the remaining 6 were normalized by the MAS5 module. Expression differences of the log2 normalized data were next statistically analyzed by a moderated t-test for micro-array data (Cyber-T) [9] and corrected for multiple testing by the Benjamini-Hochberg equation, yielding False Discovery rates (FDR) [3]. The network construction was based on the available normalized raw expression values of 1174 probes. Using a cutoff of ±2 fold change, we reduced this set to 397 differentially expressed probes. In cases where more than one probe mapped to the same gene, these probes were averaged, the average expression level was retained and 6
7 so the dataset was reduced to 315 unique genes. The mcxarray program from MCL clustering package[10] was used to construct the network from expression data. Pairwise similarities in gene expression vectors were expressed through the Pearson correlation coefficient (PCC) and a threshold of PCC>0.7 was used to link correlated genes. The resulting network representation of the gene expression experiment included proteins as nodes, edges linked correlated proteins and visualisation was performed with the Cytoscape software[11]. Transcriptomics, proteomics and metabolomics data were analysed by the MetaCoreTM systems biology analysis suite version 5.1 (GeneGo Inc., St. Joseph, MI), using a filtering criterion of p<0.05 ( Statistical analysis. Statistical analysis was performed using the analysis of variance and Student s t-test. The Kruskal-Wallis test with the Dunn s post test was used as a nonparametric test. Pairwise comparisons between metabolites were performed using the Bonferroni / Dunn test. Results were given as means±se. A P value <0.05 was considered significant. 7
8 References [1] Varghese AJ, Gulyas S, Mohindra JK. Hypoxia-dependent reduction of 1-(2- nitro-1-imidazolyl)-3-methoxy-2-propanol by Chinese hamster ovary cells and KHT tumor cells in vitro and in vivo. Cancer Res Oct; 36(10): [2] Samoszuk MK, Walter J, Mechetner E. Improved immunohistochemical method for detecting hypoxia gradients in mouse tissues and tumors. J Histochem Cytochem Jun; 52(6): [3] Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerfull approach to multiple testing. J R Statistic Soc. 1995; 57(1): [4] Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, et al. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis Nov; 21(17): [5] Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem Mar 1; 68(5): [6] Bergmeyer H. Methods of enzymatic analysis. Weinheim, Germany: Verlag Chemie; [7] May D, Gilon D, Djonov V, Itin A, Lazarus A, Gordon O, et al. Transgenic system for conditional induction and rescue of chronic myocardial hibernation provides insights into genomic programs of hibernation. Proc Natl Acad Sci U S A Jan 8; 105(1): [8] Schirmer SH, Fledderus JO, Bot PT, Moerland PD, Hoefer IE, Baan J, Jr., et al. Interferon-beta signaling is enhanced in patients with insufficient coronary collateral artery development and inhibits arteriogenesis in mice. Circ Res May 23; 102(10): [9] Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics Jun; 17(6): [10] Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for largescale detection of protein families. Nucleic Acids Res Apr 1; 30(7): [11] Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res Nov; 13(11):
9 SUPPLEMENTAL FIGURES Supplemental Figure 1. PCA and heat map. (A) PCA for multivariate analysis of proteome dynamics in hibernating (red) compared to control (blue) hearts. The black ellipse represents the 95% significance level. Principal component 1 and 2 (PC1 and PC2) clearly separate the control and hibernating hearts based on their protein expression profiles. (B) Heat map. Hierarchical clustering was applied to rearrange the dataset. X- axis, spot maps; Y-axis, proteins. Red color denotes increase, green color denotes decrease. Black color indicates no change. More proteins are upregulated than downregulated during hibernation indicating a switch in gene and protein expression. Bar shows the log standard abundance value interval for the colors, +/ Supplemental Figure 2. Changes in the proteome of hibernating hearts. The user interface of the Decyder 2D software (version 6.5, GE healthcare) displays the experimental design view for triose phosphate isomerase (spot no 42, Table 1) and creatine kinase (B, spot no 27, Table 1). Note the consistency of the differential expression in the hibernating myocardium. Supplemental Figure 3. Densitometry. Quantitative data for the phosphorylated bands of MLC-2 and TnI (highlighted with arrows in Figures 4B and C) as well as for SOD-2. *, denotes a significant difference, p<0.05 (n=4 per group) Supplemental Figure 4. Pathway analysis. The combined map (thermometer 1 is proteomics, 2 is transcriptomics, the red color indicates an upregulation in the 9
10 comparison hibernation vs control) revealed that protein changes were in the same pathway as transcriptomic changes and both datasets contributed the same number of focus molecules to the pathway analysis: Whereas proteomics identified 4 members of this pathway CCT6A (+1.25), HSPA5 (+2.18), TF (+1.32) and ALDOA (+1.52), transcriptomics identified 4 additional focus objects: EGLN3 (+6.91), BNIP (+3.2), SLC2A1 (+3.46) and VEGFA (+2.9). 10
11 Supplemental Table I: Peptides identified by LC-MS/MS No. Protein Name Accession No. Mw Protein idenumbernumbernumbe Sequence Peptide sequence Best Pepti Best SEQUBest SEQUBest X! Numbe NumbeNumb NumbCalculated + Start Stop i 1 78 kda glucose-regulated protein GRP78_MOUSE % % VEIIANDQGNR 95.00% , kda glucose-regulated protein GRP78_MOUSE % % ITPSYVAFTPEGER 95.00% , kda glucose-regulated protein GRP78_MOUSE % % NQLTSNPENTVFDAK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % TWNDPSVQQDIK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % ETAEAYLGK 95.00% kda glucose-regulated protein GRP78_MOUSE % % VTHAVVTVPAYFNDAQR 95.00% , kda glucose-regulated protein GRP78_MOUSE % % IINEPTAAAIAYGLDK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % VLEDSDLK 95.00% kda glucose-regulated protein GRP78_MOUSE % % VLEDSDLKK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % KSDIDEIVLVGGSTR 95.00% , kda glucose-regulated protein GRP78_MOUSE % % SDIDEIVLVGGSTR 95.00% , kda glucose-regulated protein GRP78_MOUSE % % IQQLVK 95.00% kda glucose-regulated protein GRP78_MOUSE % % EFFNGKEPSR 95.00% , kda glucose-regulated protein GRP78_MOUSE % % NTVVPTK 95.00% kda glucose-regulated protein GRP78_MOUSE % % NTVVPTKK 95.00% kda glucose-regulated protein GRP78_MOUSE % % SQIFSTASDNQPTVTIK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % VYEGERPLTK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % ITITNDQNR 95.00% , kda glucose-regulated protein GRP78_MOUSE % % NELESYAYSLK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % ELEEIVQPIISK 95.00% , kda glucose-regulated protein GRP78_MOUSE % % LYGSGGPPPTGEEDTSEKDEL 95.00% , Serotransferrin TRFE_MOUSE % % KGTDFQLNQLEGK 95.00% , Serotransferrin TRFE_MOUSE % % AVSSFFSGSCVPCADPVAFPK 95.00% , Serotransferrin TRFE_MOUSE % % CLKDGGGDVAFVK 95.00% , Serotransferrin TRFE_MOUSE % % DGGGDVAFVK 95.00% Serotransferrin TRFE_MOUSE % % DQYELLCLDNTR 95.00% , Serotransferrin TRFE_MOUSE % % KPVDQYEDCYLAR 95.00% , Serotransferrin TRFE_MOUSE % % SKDFQLFSSPLGK 95.00% , Serotransferrin TRFE_MOUSE % % DFQLFSSPLGK 95.00% , Serotransferrin TRFE_MOUSE % % DSAFGLLR 95.00% Serotransferrin TRFE_MOUSE % % LYLGHNYVTAIR 95.00% , Serotransferrin TRFE_MOUSE % % NQQEGVCPEGSIDNSPVK 95.00% , Serotransferrin TRFE_MOUSE % % GYYAVAVVK 95.00% Serotransferrin TRFE_MOUSE % % ASDTSITWNNLK 95.00% , Serotransferrin TRFE_MOUSE % % CLVEKGDVAFVK 95.00% , Serotransferrin TRFE_MOUSE % % NLKQEDFELLCPDGTR 95.00% , Serotransferrin TRFE_MOUSE % % WCAVSEHENTK 95.00% , Serotransferrin TRFE_MOUSE % % TSYPDCIK 95.00% Serotransferrin TRFE_MOUSE % % KGTDFQLNQLEGK 95.00% , Serotransferrin TRFE_MOUSE % % KGTDFQLNQLEGKK 95.00% , Serotransferrin TRFE_MOUSE % % GTDFQLNQLEGKK 95.00% , Serotransferrin TRFE_MOUSE % % SAGWVIPIGLLFCK 95.00% , Serotransferrin TRFE_MOUSE % % AVSSFFSGSCVPCADPVAFPK 95.00% , Serotransferrin TRFE_MOUSE % % CLKDGGGDVAFVK 95.00% , Serotransferrin TRFE_MOUSE % % DGGGDVAFVK 95.00% Serotransferrin TRFE_MOUSE % % GDVAFVK 95.00% Serotransferrin TRFE_MOUSE % % HTTIFEVLPEK 95.00% , Serotransferrin TRFE_MOUSE % % HTTIFEVLPEKADR 95.00% , Serotransferrin TRFE_MOUSE % % ADRDQYELLCLDNTR 95.00% , Serotransferrin TRFE_MOUSE % % DQYELLCLDNTR 95.00% , Serotransferrin TRFE_MOUSE % % KPVDQYEDCYLAR 95.00% , Serotransferrin TRFE_MOUSE % % IPSHAVVAR 95.00% Serotransferrin TRFE_MOUSE % % KNNGKEDLIWEILK 95.00% , Serotransferrin TRFE_MOUSE % % NNGKEDLIWEILK 95.00% , Serotransferrin TRFE_MOUSE % % VAQEHFGK 95.00% Serotransferrin TRFE_MOUSE % % SKDFQLFSSPLGK 95.00% , Serotransferrin TRFE_MOUSE % % DFQLFSSPLGK 95.00% , Serotransferrin TRFE_MOUSE % % DLLFKDSAFGLLR 95.00% , Serotransferrin TRFE_MOUSE % % DSAFGLLR 95.00% Serotransferrin TRFE_MOUSE % % LYLGHNYVTAIR 95.00% , Serotransferrin TRFE_MOUSE % % NQQEGVCPEGSIDNSPVK 95.00% , Serotransferrin TRFE_MOUSE % % WCALSHLER 95.00% , Serotransferrin TRFE_MOUSE % % TKCDEWSIISEGK 95.00% , Serotransferrin TRFE_MOUSE % % IECESAETTEDCIEK 95.00% , Serotransferrin TRFE_MOUSE % % GYYAVAVVK 95.00% Serotransferrin TRFE_MOUSE % % ASDTSITWNNLK 95.00% , Serotransferrin TRFE_MOUSE % % CAPNNKEEYNGYTGAFR 95.00% , Serotransferrin TRFE_MOUSE % % CLVEKGDVAFVK 95.00% , Serotransferrin TRFE_MOUSE % % HQTVLDNTEGKNPAEWAK 95.00% , Serotransferrin TRFE_MOUSE % % NLKQEDFELLCPDGTR 95.00% , Serotransferrin TRFE_MOUSE % % QEDFELLCPDGTR 95.00% , Serotransferrin TRFE_MOUSE % % DFASCHLAQAPNHVVVSR 95.00% ,
12 3 Serotransferrin TRFE_MOUSE % % AVLTSQETLFGGSDCTGNFCLFK 95.00% , Serotransferrin TRFE_MOUSE % % DLLFRDDTK 95.00% , Serotransferrin TRFE_MOUSE % % LPEGTTPEK 95.00% Serotransferrin TRFE_MOUSE % % LLEACTFHKH 95.00% , Serotransferrin TRFE_MOUSE % % TVLPPDGPR 95.00% Serotransferrin TRFE_MOUSE % % TSYPDCIK 95.00% Serotransferrin TRFE_MOUSE % % KGTDFQLNQLEGK 95.00% , Serotransferrin TRFE_MOUSE % % KGTDFQLNQLEGKK 95.00% , Serotransferrin TRFE_MOUSE % % DGGGDVAFVK 95.00% Serotransferrin TRFE_MOUSE % % GDVAFVK 95.00% Serotransferrin TRFE_MOUSE % % HTTIFEVLPEK 95.00% , Serotransferrin TRFE_MOUSE % % ADRDQYELLCLDNTR 95.00% , Serotransferrin TRFE_MOUSE % % DQYELLCLDNTR 95.00% , Serotransferrin TRFE_MOUSE % % KPVDQYEDCYLAR 95.00% , Serotransferrin TRFE_MOUSE % % IPSHAVVAR 95.00% Serotransferrin TRFE_MOUSE % % VAQEHFGK 95.00% Serotransferrin TRFE_MOUSE % % SKDFQLFSSPLGK 95.00% , Serotransferrin TRFE_MOUSE % % DFQLFSSPLGK 95.00% , Serotransferrin TRFE_MOUSE % % DSAFGLLR 95.00% Serotransferrin TRFE_MOUSE % % LYLGHNYVTAIR 95.00% , Serotransferrin TRFE_MOUSE % % NQQEGVCPEGSIDNSPVK 95.00% , Serotransferrin TRFE_MOUSE % % TKCDEWSIISEGK 95.00% , Serotransferrin TRFE_MOUSE % % IECESAETTEDCIEK 95.00% , Serotransferrin TRFE_MOUSE % % GYYAVAVVK 95.00% Serotransferrin TRFE_MOUSE % % ASDTSITWNNLK 95.00% , Serotransferrin TRFE_MOUSE % % FDEFFSQGCAPGYEK 95.00% , Serotransferrin TRFE_MOUSE % % CAPNNKEEYNGYTGAFR 95.00% , Serotransferrin TRFE_MOUSE % % EEYNGYTGAFR 95.00% , Serotransferrin TRFE_MOUSE % % CLVEKGDVAFVK 95.00% , Serotransferrin TRFE_MOUSE % % HQTVLDNTEGK 95.00% , Serotransferrin TRFE_MOUSE % % HQTVLDNTEGKNPAEWAK 95.00% , Serotransferrin TRFE_MOUSE % % NPAEWAK 95.00% Serotransferrin TRFE_MOUSE % % NLKQEDFELLCPDGTR 95.00% , Serotransferrin TRFE_MOUSE % % QEDFELLCPDGTR 95.00% , Serotransferrin TRFE_MOUSE % % DFASCHLAQAPNHVVVSR 95.00% , Serotransferrin TRFE_MOUSE % % DLLFRDDTK 95.00% , Serotransferrin TRFE_MOUSE % % LPEGTTPEK 95.00% Serotransferrin TRFE_MOUSE % % LLEACTFHK 95.00% , Aconitate hydratase, mitochondrial ACON_MOUSE % % VAGILTVK 95.00% Aconitate hydratase, mitochondrial ACON_MOUSE % % SQFTITPGSEQIR 95.00% , Aconitate hydratase, mitochondrial ACON_MOUSE % % DGYAQILR 95.00% Aconitate hydratase, mitochondrial ACON_MOUSE % % NTIVTSYNR 95.00% , Aconitate hydratase, mitochondrial ACON_MOUSE % % VDVSPTSQR 95.00% Aconitate hydratase, mitochondrial ACON_MOUSE % % NAVTQEFGPVPDTAR 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % LPFSIR 95.00% Cytoplasmic aconitate hydratase ACOC_MOUSE % % NCDEFLVK 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % NIEVPFKPAR 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % NQDLEFER 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % DFESCLGAK 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % GFQVAPDR 95.00% Cytoplasmic aconitate hydratase ACOC_MOUSE % % TSLSPGSGVVTYYLR 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % IDFEKEPLGVNAQGR 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % SWNALAAPSEK 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % LYAWNPK 95.00% Cytoplasmic aconitate hydratase ACOC_MOUSE % % YQQAGLPLIVLAGK 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % AVLAESYER 95.00% , Cytoplasmic aconitate hydratase ACOC_MOUSE % % YTINIPEDLKPR 95.00% , Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % IIQVAK 95.00% Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % EFQEQLESAR 95.00% , Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % IIEEAPAPGINPEVR 95.00% , Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % LGEAAVR 95.00% Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % LVVWASDR 95.00% Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % SNVDFLLR 95.00% Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % DLLPSHSTIAK 95.00% , Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % RLNISYTR 95.00% , Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % LNISYTR 95.00% Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % VLGDLSSEDGCTYLK 95.00% , Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial MCCA_MOUSE % % VFFSEGAQANR 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % GLQDVLR 95.00% T-complex protein 1 subunit zeta TCPZ_MOUSE % % QADLYISEGLHPR 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % IITEGFEAAK 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % ALQFLEQVK 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % ETLIDVAR 95.00% T-complex protein 1 subunit zeta TCPZ_MOUSE % % SETDTSLIR 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % TEVNSGFFYK 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % GFVVINQK 95.00%
13 8 T-complex protein 1 subunit zeta TCPZ_MOUSE % % GIDPFSLDALAK 95.00% , T-complex protein 1 subunit zeta TCPZ_MOUSE % % VLAQNSGFDLQETLVK 95.00% , Protein disulfide-isomerase PDIA1_MOUSE % % NNFEGEITK 95.00% , Protein disulfide-isomerase PDIA1_MOUSE % % LLDFIK 95.00% Protein disulfide-isomerase PDIA1_MOUSE % % YKPESDELTAEK 95.00% , Protein disulfide-isomerase PDIA1_MOUSE % % QLAPIWDK 95.00% Protein disulfide-isomerase PDIA1_MOUSE % % TVIDYNGER 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % EIVHLQAGQCGNQIGAK 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % INVYYNEATGGK 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % FPGQLNADLR 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % FPGQLNADLRK 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % LAVNMVPFPR 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % YLTVAAVFR 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % NSSYFVEWIPNNVK 95.00% , Tubulin beta-2c chain TBB2C_MOUSE % % TAVCDIPPR 95.00% , Desmin DESM_MOUSE % % VYQVSR 95.00% Desmin DESM_MOUSE % % TSGGAGGLGSLR 95.00% , Desmin DESM_MOUSE % % TNEKVELQELNDR 95.00% , Desmin DESM_MOUSE % % VELQELNDR 95.00% , Desmin DESM_MOUSE % % FANYIEK 95.00% Desmin DESM_MOUSE % % FLEQQNAALAAEVNR 95.00% , Desmin DESM_MOUSE % % VAELYEEEMR 95.00% , Desmin DESM_MOUSE % % QVEVLTNQR 95.00% , Desmin DESM_MOUSE % % VDVERDNLIDDLQR 95.00% , Desmin DESM_MOUSE % % DNLIDDLQR 95.00% , Desmin DESM_MOUSE % % LQEEIQLR 95.00% , Desmin DESM_MOUSE % % EEAENNLAAFR 95.00% , Desmin DESM_MOUSE % % ADVDAATLAR 95.00% , Desmin DESM_MOUSE % % IESLNEEIAFLK 95.00% , Desmin DESM_MOUSE % % AQYETIAAK 95.00% Desmin DESM_MOUSE % % NISEAEEWYK 95.00% , Desmin DESM_MOUSE % % VSDLTQAANK 95.00% , Desmin DESM_MOUSE % % VSDLTQAANKNNDALR 95.00% , Desmin DESM_MOUSE % % NNDALR 95.00% Desmin DESM_MOUSE % % HQIQSYTCEIDALK 95.00% , Desmin DESM_MOUSE % % FASEANGYQDNIAR 95.00% , Desmin DESM_MOUSE % % EYQDLLNVK 95.00% , Desmin DESM_MOUSE % % KLLEGEESR 95.00% , Desmin DESM_MOUSE % % LLEGEESR 95.00% Desmin DESM_MOUSE % % INLPIQTFSALNFR 95.00% , Desmin DESM_MOUSE % % DGEVVSEATQQQHEVL 95.00% , Desmin DESM_MOUSE % % TSGGAGGLGSLR 95.00% , Desmin DESM_MOUSE % % TNEKVELQELNDR 95.00% , Desmin DESM_MOUSE % % VELQELNDR 95.00% , Desmin DESM_MOUSE % % FANYIEK 95.00% Desmin DESM_MOUSE % % FLEQQNAALAAEVNR 95.00% , Desmin DESM_MOUSE % % VAELYEEEMR 95.00% , Desmin DESM_MOUSE % % RQVEVLTNQR 95.00% , Desmin DESM_MOUSE % % QVEVLTNQR 95.00% , Desmin DESM_MOUSE % % VDVERDNLIDDLQR 95.00% , Desmin DESM_MOUSE % % DNLIDDLQR 95.00% , Desmin DESM_MOUSE % % LQEEIQLR 95.00% , Desmin DESM_MOUSE % % EEAENNLAAFR 95.00% , Desmin DESM_MOUSE % % ADVDAATLAR 95.00% , Desmin DESM_MOUSE % % RIESLNEEIAFLKK 95.00% , Desmin DESM_MOUSE % % IESLNEEIAFLK 95.00% , Desmin DESM_MOUSE % % VHEEEIR 95.00% Desmin DESM_MOUSE % % AQYETIAAK 95.00% Desmin DESM_MOUSE % % NISEAEEWYK 95.00% , Desmin DESM_MOUSE % % VSDLTQAANK 95.00% , Desmin DESM_MOUSE % % VSDLTQAANKNNDALR 95.00% , Desmin DESM_MOUSE % % HQIQSYTCEIDALK 95.00% , Desmin DESM_MOUSE % % FASEANGYQDNIAR 95.00% , Desmin DESM_MOUSE % % EYQDLLNVK 95.00% , Desmin DESM_MOUSE % % KLLEGEESR 95.00% , Desmin DESM_MOUSE % % LLEGEESR 95.00% Desmin DESM_MOUSE % % INLPIQTFSALNFR 95.00% , Desmin DESM_MOUSE % % DGEVVSEATQQQHEVL 95.00% , Fibrinogen gamma chain FIBG_MOUSE % % DNCCILDER 95.00% , Fibrinogen gamma chain FIBG_MOUSE % % TLEDILFR 95.00% , Fibrinogen gamma chain FIBG_MOUSE % % YLQEIYNSNNQK 95.00% , Protein disulfide-isomerase A3 PDIA3_MOUSE % % LAPEYEAAATR 95.00% , Protein disulfide-isomerase A3 PDIA3_MOUSE % % YGVSGYPTLK 95.00% , Protein disulfide-isomerase A3 PDIA3_MOUSE % % QAGPASVPLR 95.00% Protein disulfide-isomerase A3 PDIA3_MOUSE % % LNFAVASR 95.00% Protein disulfide-isomerase A3 PDIA3_MOUSE % % SEPIPESNEGPVK 95.00% ,
14 14 Protein disulfide-isomerase A3 PDIA3_MOUSE % % DPNIVIAK 95.00% Fibrinogen beta chain FIBB_MOUSE % % SILEDLR 95.00% Fibrinogen beta chain FIBB_MOUSE % % TPCTVSCNIPVVSGK 95.00% , Fibrinogen beta chain FIBB_MOUSE % % TENGGWTVIQNR 95.00% , Fibrinogen beta chain FIBB_MOUSE % % QDGSVDFGR 95.00% Fibrinogen beta chain FIBB_MOUSE % % GFGNIATNEDAK 95.00% , Fibrinogen beta chain FIBB_MOUSE % % ISQLTR 95.00% Fibrinogen beta chain FIBB_MOUSE % % AHYGGFTVQNEASK 95.00% , Fibrinogen beta chain FIBB_MOUSE % % YQVSVNK 95.00% Fibrinogen beta chain FIBB_MOUSE % % DNDGWVTTDPR 95.00% , Fibrinogen beta chain FIBB_MOUSE % % IRPFFPQQ 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % LDIDSAPITAR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % NTGIICTIGPASR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % EATESFASDPILYRPVAVALDTK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % PVAVALDTK 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % TGLIKGSGTAEVELKK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GSGTAEVELKK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % CDENILWLDYK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % CDENILWLDYKNICK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % IYVDDGLISLQVK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % EKGADFLVTEVENGGSLGSK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % EKGADFLVTEVENGGSLGSKK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GADFLVTEVENGGSLGSK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GADFLVTEVENGGSLGSKK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % KGVNLPGAAVDLPAVSEKDIQDLK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GVNLPGAAVDLPAVSEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GVNLPGAAVDLPAVSEKDIQDLK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDLGIEIPAEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDLGIEIPAEKVFLAQK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDYPLEAVR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % APIIAVTR 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GIFPVLCK 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % DAVLNAWAEDVDLR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % VNLAMDVGK 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % LDIDSAPITAR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % NTGIICTIGPASR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GSGTAEVELKK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % CDENILWLDYK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % VVEVGSK 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % EKGADFLVTEVENGGSLGSK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GADFLVTEVENGGSLGSK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GADFLVTEVENGGSLGSKK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % KGVNLPGAAVDLPAVSEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GVNLPGAAVDLPAVSEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GVNLPGAAVDLPAVSEKDIQDLK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % KAADVHEVR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % AADVHEVR 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % IENHEGVR 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDLGIEIPAEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDYPLEAVR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % APIIAVTR 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GIFPVLCK 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % VNLAMDVGK 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % LDIDSAPITAR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % NTGIICTIGPASR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % CDENILWLDYK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GADFLVTEVENGGSLGSK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GVNLPGAAVDLPAVSEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDLGIEIPAEK 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GDYPLEAVR 95.00% , Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % APIIAVTR 95.00% Pyruvate kinase isozymes M1/M2 KPYM_MOUSE % % GIFPVLCK 95.00% Cytosol aminopeptidase AMPL_MOUSE % % DKDDDLPQFTSAGESFNK 95.00% , Cytosol aminopeptidase AMPL_MOUSE % % GVLFASGQNLAR 95.00% , Cytosol aminopeptidase AMPL_MOUSE % % TIQVDNTDAEGR 95.00% , Cytosol aminopeptidase AMPL_MOUSE % % QVIDCQLADVNNLGK 95.00% , Cytosol aminopeptidase AMPL_MOUSE % % SAGACTAAAFLR 95.00% , Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % FYTDPVEAVK 95.00% , Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % MISSYVGENAEFER 95.00% , Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % AGGAGVPAFYTSTGYGTLVQEGGSPIK 95.00% , Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % DGSVAIASKPR 95.00% , Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % AGNVIFR 95.00% Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % DEADADLINAGK 95.00% , Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % CTLPLTGK 95.00% Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial SCOT1_MOUSE % % GVFDVDKK 95.00% Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % GASIVEDK 95.00%
15 20 Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % GASIVEDKLVEDLK 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % LVEDLK 95.00% Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % DDGSWEVIEGYR 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % YSTDVSVDEVK 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % CAVVDVPFGGAK 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % PISQGGIHGR 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % ELEDFK 95.00% Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % LQHGSILGFPK 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % IIAEGANGPTTPEADKIFLER 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % NLNHVSYGR 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % HGGTIPVVPTAEFQDR 95.00% , Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % YNLGLDLR 95.00% Glutamate dehydrogenase 1, mitochondrial DHE3_MOUSE % % TAAYVNAIEK 95.00% , Beta-enolase ENOB_MOUSE % % TLGPALLEK 95.00% Beta-enolase ENOB_MOUSE % % TAIQAAGYPDK 95.00% , Beta-enolase ENOB_MOUSE % % YDLDFK 95.00% Beta-enolase ENOB_MOUSE % % LGELYK 95.00% Beta-enolase ENOB_MOUSE % % ACNCLLLK 95.00% Beta-enolase ENOB_MOUSE % % VNQIGSVTESIQACK 95.00% , Beta-enolase ENOB_MOUSE % % IEEALGDK 95.00% Adenosine kinase ADK_MOUSE % % FGEILK 95.00% Adenosine kinase ADK_MOUSE % % TVIFTQGR 95.00% Ornithine aminotransferase, mitochondrial OAT_MOUSE % % IADEIQTGLAR 95.00% , Ornithine aminotransferase, mitochondrial OAT_MOUSE % % LPSDVVTSVR 95.00% , Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % LEEGPPVTTVLTR 95.00% , Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % ADQLYK 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % AHGFTFTR 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % AILAELTGR 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % GAGIALACK 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % LPCIFICENNR 95.00% , Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % AAASTDYYK 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % GDFIPGLR 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % VDGMDILCVR 95.00% , Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % EATKFAAAYCR 95.00% , Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % FAAAYCR 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % TREEIQEVR 95.00% , Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % EEIQEVR 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % EIDVEVR 95.00% Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial ODPA_MOUSE % % GANQWIK 95.00% Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % QGLLGINIAEK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % SGSDWILNGSK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LNYKPQEEYPDLSK 95.00% , Creatine kinase M-type KCRM_MOUSE % % VLTPDLYNKLR 95.00% , Creatine kinase M-type KCRM_MOUSE % % DLFDPIIQDR 95.00% , Creatine kinase M-type KCRM_MOUSE % % TDLNHENLKGGDDLDPNYVLSSR 95.00% , Creatine kinase M-type KCRM_MOUSE % % GGDDLDPNYVLSSR 95.00% , Creatine kinase M-type KCRM_MOUSE % % GYTLPPHCSR 95.00% , Creatine kinase M-type KCRM_MOUSE % % AVEKLSVEALNSLTGEFK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LSVEALNSLTGEFK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LSVEALNSLTGEFKGK 95.00% , Creatine kinase M-type KCRM_MOUSE % % YYPLK 95.00% Creatine kinase M-type KCRM_MOUSE % % DWPDAR 95.00% Creatine kinase M-type KCRM_MOUSE % % SFLVWVNEEDHLR 95.00% , Creatine kinase M-type KCRM_MOUSE % % RFCVGLQK 95.00% , Creatine kinase M-type KCRM_MOUSE % % FCVGLQK 95.00% Creatine kinase M-type KCRM_MOUSE % % LANLSK 95.00% Creatine kinase M-type KCRM_MOUSE % % HPKFEEILTR 95.00% , Creatine kinase M-type KCRM_MOUSE % % FEEILTR 95.00% Creatine kinase M-type KCRM_MOUSE % % RGTGGVDTAAVGAVFDISNADR 95.00% , Creatine kinase M-type KCRM_MOUSE % % GTGGVDTAAVGAVFDISNADR 95.00% , Creatine kinase M-type KCRM_MOUSE % % LGSSEVEQVQLVVDGVK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LGSSEVEQVQLVVDGVKLMVEMEK 95.00% , Creatine kinase M-type KCRM_MOUSE % % GQSIDDMIPAQK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % RIFSSEHDIFR 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % IFSSEHDIFR 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % FFQEEVIPHHTEWEK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % AGEVSREVWEK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % AGKQGLLGINIAEK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % QGLLGINIAEK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % CIGAIAMTEPGAGSDLQGVR 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % RSGSDWILNGSK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % SGSDWILNGSK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % SPAHGISLFLVENGMK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % AQDTAELFFEDVR 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % LPANALLGEENK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % GFYYLMQELPQER 95.00% ,
16 26 Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % THICVTR 95.00% Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % AFVDSCLQLHETK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % AFVDSCLQLHETKR 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % VQPIYGGTNEIMK 95.00% , Long-chain specific acyl-coa dehydrogenase, mitochondrial ACADL_MOUSE % % VQPIYGGTNEIMKELIAR 95.00% , Creatine kinase M-type KCRM_MOUSE % % FKLNYKPQEEYPDLSK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LNYKPQEEYPDLSK 95.00% , Creatine kinase M-type KCRM_MOUSE % % VLTPDLYNK 95.00% , Creatine kinase M-type KCRM_MOUSE % % DLFDPIIQDR 95.00% , Creatine kinase M-type KCRM_MOUSE % % HGGYKPTDK 95.00% , Creatine kinase M-type KCRM_MOUSE % % TDLNHENLK 95.00% , Creatine kinase M-type KCRM_MOUSE % % TDLNHENLKGGDDLDPNYVLSSR 95.00% , Creatine kinase M-type KCRM_MOUSE % % GGDDLDPNYVLSSR 95.00% , Creatine kinase M-type KCRM_MOUSE % % GYTLPPHCSR 95.00% , Creatine kinase M-type KCRM_MOUSE % % LSVEALNSLTGEFK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LSVEALNSLTGEFKGK 95.00% , Creatine kinase M-type KCRM_MOUSE % % YYPLK 95.00% Creatine kinase M-type KCRM_MOUSE % % SFLVWVNEEDHLR 95.00% , Creatine kinase M-type KCRM_MOUSE % % RFCVGLQK 95.00% , Creatine kinase M-type KCRM_MOUSE % % FCVGLQK 95.00% Creatine kinase M-type KCRM_MOUSE % % VLTCPSNLGTGLR 95.00% , Creatine kinase M-type KCRM_MOUSE % % LANLSK 95.00% Creatine kinase M-type KCRM_MOUSE % % HPKFEEILTR 95.00% , Creatine kinase M-type KCRM_MOUSE % % FEEILTR 95.00% Creatine kinase M-type KCRM_MOUSE % % RGTGGVDTAAVGAVFDISNADR 95.00% , Creatine kinase M-type KCRM_MOUSE % % GTGGVDTAAVGAVFDISNADR 95.00% , Creatine kinase M-type KCRM_MOUSE % % LGSSEVEQVQLVVDGVK 95.00% , Creatine kinase M-type KCRM_MOUSE % % LGSSEVEQVQLVVDGVKLMVEMEK 95.00% , Creatine kinase M-type KCRM_MOUSE % % GQSIDDMIPAQK 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % KELSDIAHR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % ELSDIAHR 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % GILAADESTGSIAK 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % GILAADESTGSIAKR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % RLQSIGTENTEENRR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % LQSIGTENTEENR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % QLLLTADDR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % ADDGRPFPQVIK 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % GGVVGIKVDK 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % VDKGVVPLAGTNGETTTQGLDGLSER 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % GVVPLAGTNGETTTQGLDGLSER 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % DGADFAK 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % YASICQQNGIVPIVEPEILPDGDHDLK 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % YASICQQNGIVPIVEPEILPDGDHDLKR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % CQYVTEK 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % VLAAVYK 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % ALQASALK 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % AAQEEYIK 95.00% Fructose-bisphosphate aldolase A ALDOA_MOUSE % % AAQEEYIKR 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % RALANSLACQGK 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % ALANSLACQGK 95.00% , Fructose-bisphosphate aldolase A ALDOA_MOUSE % % YTPSGQSGAAASESLFISNHAY 95.00% , Aldose reductase ALDR_MOUSE % % SPPGQVTEAVK 95.00% , Aldose reductase ALDR_MOUSE % % VAIDLGYR 95.00% Aldose reductase ALDR_MOUSE % % HIDCAQVYQNEK 95.00% , Aldose reductase ALDR_MOUSE % % EVGVALQEK 95.00% Aldose reductase ALDR_MOUSE % % QDLFIVSK 95.00% Aldose reductase ALDR_MOUSE % % LWCTFHDK 95.00% , Aldose reductase ALDR_MOUSE % % TIGVSNFNPLQIER 95.00% , Aldose reductase ALDR_MOUSE % % ILNKPGLK 95.00% Aldose reductase ALDR_MOUSE % % LIEYCHSK 95.00% , Aldose reductase ALDR_MOUSE % % PEDPSLLEDPR 95.00% , Aldose reductase ALDR_MOUSE % % TTAQVLIR 95.00% Aldose reductase ALDR_MOUSE % % NLVVIPK 95.00% Aldose reductase ALDR_MOUSE % % HKDYPFHAEV 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % LVINGKPITIFQER 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % IVSNASCTTNCLAPLAK 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % DGRGAAQNIIPASTGAAK 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % GAAQNIIPASTGAAK 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % VIPELNGK 95.00% Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % VPTPNVSVVDLTCR 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % LISWYDNEYGYSNR 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % VVDLMAYMASKE 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % VFVTGPLPAEGR 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % KDLEQGVVGAHGLLCR 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % DLEQGVVGAHGLLCR 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % KLLDAAGANLR 95.00% ,
17 30 Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % LLDAAGANLR 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % RLPEAIEEVK 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % LPEAIEEVK 95.00% , Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % LKPFGVQR 95.00% Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % FLYTGR 95.00% Glyoxylate reductase/hydroxypyruvate reductase GRHPR_MOUSE % % NCVILPHIGSATYK 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % VGVNGFGR 95.00% Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % AAICSGK 95.00% Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % LVINGKPITIFQER 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % PITIFQER 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % IVSNASCTTNCLAPLAK 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % GAAQNIIPASTGAAK 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % LTGMAFRVPTPNVSVVDLTCR 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % VPTPNVSVVDLTCR 95.00% , Glyceraldehyde-3-phosphate dehydrogenase G3P_MOUSE % % LISWYDNEYGYSNR 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % LVIITAGAR 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % LNLVQR 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % NVNIFK 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % FIIPNIVK 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % QVVDSAYEVIK 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % VTLTPEEEAR 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % DQLIVNLLK 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % DYCVTANSK 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % LVIITAGAR 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % LNLVQR 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % NVNIFK 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % ISGFPK 95.00% L-lactate dehydrogenase A chain LDHA_MOUSE % % VIGSGCNLDSAR 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % SLNPELGTDADK 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % QVVDSAYEVIK 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % VTLTPEEEAR 95.00% , L-lactate dehydrogenase A chain LDHA_MOUSE % % SADTLWGIQK 95.00% , Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial ECH1_MOUSE % % ELVECFQK 95.00% , Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial ECH1_MOUSE % % AVVVSGAGK 95.00% Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial ECH1_MOUSE % % SPVAVQGSK 95.00% Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial ECH1_MOUSE % % INLIYSR 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % KGIEESLK 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % KGIEESLKR 95.00% , Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % GIEESLKR 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % AGDEFVEK 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % LKNELFQR 95.00% , Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % NELFQR 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % LVEVIK 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % TFESLVDFCK 95.00% , Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % DTPGFIVNR 95.00% , Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % LLVPYLIEAVR 95.00% , Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % KTGEGFYK 95.00% Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial HCDH_MOUSE % % TGEGFYK 95.00% ATP synthase subunit gamma, mitochondrial ATPG_MOUSE % % HLIIGVSSDR 95.00% , ATP synthase subunit gamma, mitochondrial ATPG_MOUSE % % QMKNEVAALTAAGK 95.00% , ATP synthase subunit gamma, mitochondrial ATPG_MOUSE % % NEVAALTAAGK 95.00% , ATP synthase subunit gamma, mitochondrial ATPG_MOUSE % % EVMIVGVGEK 95.00% , ATP synthase subunit gamma, mitochondrial ATPG_MOUSE % % THSDQFLVSFK 95.00% , Troponin I, cardiac muscle TNNI3_MOUSE % % AYATEPHAK 95.00% Troponin I, cardiac muscle TNNI3_MOUSE % % NITEIADLTQK 95.00% , Apolipoprotein A-I APOA1_MOUSE % % LGPLTR 95.00% Apolipoprotein A-I APOA1_MOUSE % % VQPYLDEFQK 95.00% , Apolipoprotein A-I APOA1_MOUSE % % EDVELYR 95.00% Apolipoprotein A-I APOA1_MOUSE % % VAPLGAELQESAR 95.00% , Apolipoprotein A-I APOA1_MOUSE % % LSPVAEEFR 95.00% , Apolipoprotein A-I APOA1_MOUSE % % PALEDLR 95.00% Heat shock protein beta-1 HSPB1_MOUSE % % LFDQAFGVPR 95.00% , Heat shock protein beta-1 HSPB1_MOUSE % % EGVVEITGK 95.00% Triosephosphate isomerase TPIS_MOUSE % % FFVGGNWK 95.00% Triosephosphate isomerase TPIS_MOUSE % % IAVAAQNCYK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % HVFGESDELIGQK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % VVFEQTK 95.00% Triosephosphate isomerase TPIS_MOUSE % % IIYGGSVTGATCK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % KFFVGGNWK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % FFVGGNWK 95.00% Triosephosphate isomerase TPIS_MOUSE % % IAVAAQNCYK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % VTNGAFTGEISPGMIK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % DLGATWVVLGHSER 95.00% , Triosephosphate isomerase TPIS_MOUSE % % RHVFGESDELIGQK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % HVFGESDELIGQK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % LDEREAGITEK 95.00% ,
18 41 Triosephosphate isomerase TPIS_MOUSE % % EAGITEK 95.00% Triosephosphate isomerase TPIS_MOUSE % % VVFEQTK 95.00% Triosephosphate isomerase TPIS_MOUSE % % VIADNVK 95.00% Triosephosphate isomerase TPIS_MOUSE % % VIADNVKDWSK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % VVLAYEPVWAIGTGK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % TATPQQAQEVHEK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % SNVNDGVAQSTR 95.00% , Triosephosphate isomerase TPIS_MOUSE % % IIYGGSVTGATCK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % FFVGGNWK 95.00% Triosephosphate isomerase TPIS_MOUSE % % IAVAAQNCYK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % VTNGAFTGEISPGMIK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % HVFGESDELIGQK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % LDEREAGITEK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % VVFEQTK 95.00% Triosephosphate isomerase TPIS_MOUSE % % VIADNVK 95.00% Triosephosphate isomerase TPIS_MOUSE % % VIADNVKDWSK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % VVLAYEPVWAIGTGK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % TATPQQAQEVHEK 95.00% , Triosephosphate isomerase TPIS_MOUSE % % SNVNDGVAQSTR 95.00% , Triosephosphate isomerase TPIS_MOUSE % % IIYGGSVTGATCK 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % EAFTIMDQNR 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % DGFIDK 95.00% Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % DGFIDKNDLR 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % DTFAALGR 95.00% Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % NEEIDEMIK 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % LKGADPEETILNAFK 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % GADPEETILNAFK 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % VFDPEGK 95.00% Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % ADYVR 95.00% Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % NLVHIITHGEEK 95.00% , Myosin regulatory light chain 2, ventricular/cardiac muscle isoform MLRV_MOUSE % % NLVHIITHGEEKD 95.00% , Peroxiredoxin-2 PRDX2_MOUSE % % LSDYR 95.00% Peroxiredoxin-2 PRDX2_MOUSE % % SLSQNYGVLK 95.00% , Peroxiredoxin-2 PRDX2_MOUSE % % NDEGIAYR 95.00% Peroxiredoxin-2 PRDX2_MOUSE % % GLFIIDAK 95.00% Peroxiredoxin-2 PRDX2_MOUSE % % QITVNDLPVGR 95.00% , Peroxiredoxin-2 PRDX2_MOUSE % % SVDEALR 95.00% Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % TVYHAEEIQCNGR 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % ALDSTTVAAHESEIYCK 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % NFGPTGIGFGGLTQQVEK 95.00% , NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % TPAPSPQTSLPNPITYLTK 95.00% , NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % AYDLVVDWPVTLVR 95.00% , NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % TYYYHR 95.00% NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % RVPDITECK 95.00% , NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % VPDITECK 95.00% NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % ELEQFTK 95.00% NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % AYQDRYLDLGAYYSAR 95.00% , NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 NDUBA_MOUSE % % YLDLGAYYSAR 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % TVYHAEEIQCNGR 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % ALDSTTVAAHESEIYCK 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % FGESEKCPR 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % SLESTNVTDKDGELYCK 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % NFGPTGIGFGGLTQQVEK 95.00% , Cysteine and glycine-rich protein 3 CSRP3_MOUSE % % NFGPTGIGFGGLTQQVEKKE 95.00% , Glutathione S-transferase P 1 GSTP1_MOUSE % % FEDGDLTLYQSNAILR 95.00% , Glutathione S-transferase P 1 GSTP1_MOUSE % % SLGLYGK 95.00% Glutathione S-transferase P 1 GSTP1_MOUSE % % YVTLIYTNYENGKNDYVK 95.00% , Glutathione S-transferase P 1 GSTP1_MOUSE % % ALPGHLKPFETLLSQNQGGK 95.00% , Glutathione S-transferase P 1 GSTP1_MOUSE % % AFLSSPEHVNRPINGNGK 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % RPFFPFHSPSR 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % APSWIDTGLSEMR 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % HFSPEELK 95.00% Alpha-crystallin B chain CRYAB_MOUSE % % HFSPEELKVK 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % VLGDVIEVHGK 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % QDEHGFISR 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % KQVSGPER 95.00% Alpha-crystallin B chain CRYAB_MOUSE % % TIPITR 95.00% Alpha-crystallin B chain CRYAB_MOUSE % % TIPITREEKPAVAAAPK 95.00% , Alpha-crystallin B chain CRYAB_MOUSE % % EEKPAVAAAPK 95.00% , Peroxiredoxin-1 PRDX1_MOUSE % % IGYPAPNFK 95.00% , Peroxiredoxin-1 PRDX1_MOUSE % % DISLSEYK 95.00% Peroxiredoxin-1 PRDX1_MOUSE % % DISLSEYKGK 95.00% , Peroxiredoxin-1 PRDX1_MOUSE % % TIAQDYGVLK 95.00% , Peroxiredoxin-1 PRDX1_MOUSE % % ADEGISFR 95.00% Peroxiredoxin-1 PRDX1_MOUSE % % GLFIIDDK 95.00% Peroxiredoxin-1 PRDX1_MOUSE % % GLFIIDDKGILR 95.00% ,
19 48 Peroxiredoxin-1 PRDX1_MOUSE % % QITINDLPVGR 95.00% , Peroxiredoxin-1 PRDX1_MOUSE % % SVDEIIR 95.00% Peroxiredoxin-1 PRDX1_MOUSE % % LVQAFQFTDK 95.00% , Peroxiredoxin-1 PRDX1_MOUSE % % SKEYFSK 95.00% Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % AVILGPPGSGK 95.00% Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % AVILGPPGSGKGTVCER 95.00% , Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % GLLVPDHVITR 95.00% , Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % SAQHWLLDGFPR 95.00% , Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % YKDAAKPVIELYK 95.00% , Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % DAAKPVIELYK 95.00% , Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % GVLHQFSGTETNR 95.00% , Adenylate kinase isoenzyme 4, mitochondrial KAD4_MOUSE % % IWPYVYTLFSNK 95.00% ,
20 Supplemental Table II. Overlap of significant protein and mrna changes only Protein identity Gene Symbol Gene Name PROTEOMICS Fold P-value change TRANSCRIPTOMICS Fold Bayes. FDR change p Adenylate kinase isoenzyme 4, AK3L1 Adenylate kinase 3, alpha-like mitochondrial Short chain 3-hydroxyacyl- HADHSC L-3_Hydroxyacyl-CoA CoA dehydrogenase, mitochondrial dehydrogenase, short chain L-lactate dehydrogenase A LDHA Lactate dehydrogenase A E Methylcrotonoyl-CoA MCCC1 Methylcrotonyl-CoA carboxylase carboxylase alpha chain, mitochondrial 1, alpha Succinyl-CoA:3-ketoacidcoenzyme OXCT1 3-Oxoacid CoA transferase A transferase 1, mitochondrial Protein disulfide-isomerase P4HB Prolyl 4-Hydroxylase, beta Pyruvate kinase, isozyme M2 PKM2 Pyruvate kinase, isozyme M E Triosephosphate isomerase TPI1 Triosephosphate isomerase E Serotransferrin TRF Transferrin
21 Supplemental Table III: Pathway identification using Metacore TM Supp p-value: Metacore statistics for pathway identification, Objects: genes/proteins/metabolites identified in map
22 Supplemental Figure 1 Mayr et al A B
23 Supplemental Figure 2 Mayr et al Spot 42: Triose phosphate isomerase fold P=1.4e-008 Spot 27: Creatine Kinase M fold P=0.0018
24 Supplemental Figure 3 Mayr et al p=0.023 p=0.012 p=0.015
25 Supplemental Figure 4 Mayr et al
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic acids with the assistance of isocyanide
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
ΠΡΩΤΕΩΜΙΚΗ : ΤΕΧΝΟΛΟΓΙΕΣ ΚΑΙ ΕΦΑΡΜΟΓΕΣ. Γεώργιος Θ. Τσάγκαρης, PhD Ερευνητική Μονάδα Πρωτεωµικής
 ΠΡΩΤΕΩΜΙΚΗ : ΤΕΧΝΟΛΟΓΙΕΣ ΚΑΙ ΕΦΑΡΜΟΓΕΣ Γεώργιος Θ. Τσάγκαρης, PhD Ερευνητική Μονάδα Πρωτεωµικής Γένωµα v/s Πρωτέωµα κ.λ.π. Γέν-ωµα (γίνοµαι + -ωµα) Μεταγράφηµα (transcriptome) Πρωτέ-ωµα (πρωτεΐνη + -ωµα)
ΠΡΩΤΕΩΜΙΚΗ : ΤΕΧΝΟΛΟΓΙΕΣ ΚΑΙ ΕΦΑΡΜΟΓΕΣ Γεώργιος Θ. Τσάγκαρης, PhD Ερευνητική Μονάδα Πρωτεωµικής Γένωµα v/s Πρωτέωµα κ.λ.π. Γέν-ωµα (γίνοµαι + -ωµα) Μεταγράφηµα (transcriptome) Πρωτέ-ωµα (πρωτεΐνη + -ωµα)
ΖΩΝΟΠΟΙΗΣΗ ΤΗΣ ΚΑΤΟΛΙΣΘΗΤΙΚΗΣ ΕΠΙΚΙΝΔΥΝΟΤΗΤΑΣ ΣΤΟ ΟΡΟΣ ΠΗΛΙΟ ΜΕ ΤΗ ΣΥΜΒΟΛΗ ΔΕΔΟΜΕΝΩΝ ΣΥΜΒΟΛΟΜΕΤΡΙΑΣ ΜΟΝΙΜΩΝ ΣΚΕΔΑΣΤΩΝ
 EΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΕΙΟ Τμήμα Μηχανικών Μεταλλείων-Μεταλλουργών ΖΩΝΟΠΟΙΗΣΗ ΤΗΣ ΚΑΤΟΛΙΣΘΗΤΙΚΗΣ ΕΠΙΚΙΝΔΥΝΟΤΗΤΑΣ ΜΕ ΤΗ ΣΥΜΒΟΛΗ ΔΕΔΟΜΕΝΩΝ ΣΥΜΒΟΛΟΜΕΤΡΙΑΣ ΜΟΝΙΜΩΝ ΣΚΕΔΑΣΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Κιτσάκη Μαρίνα
EΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΕΙΟ Τμήμα Μηχανικών Μεταλλείων-Μεταλλουργών ΖΩΝΟΠΟΙΗΣΗ ΤΗΣ ΚΑΤΟΛΙΣΘΗΤΙΚΗΣ ΕΠΙΚΙΝΔΥΝΟΤΗΤΑΣ ΜΕ ΤΗ ΣΥΜΒΟΛΗ ΔΕΔΟΜΕΝΩΝ ΣΥΜΒΟΛΟΜΕΤΡΙΑΣ ΜΟΝΙΜΩΝ ΣΚΕΔΑΣΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Κιτσάκη Μαρίνα
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Supporting Information
 Supporting Information Wiley-VC 007 9 Weinheim, Germany ew ear Infrared Dyes and Fluorophores Based on Diketopyrrolopyrroles Dipl.-Chem. Georg M. Fischer, Dipl.-Chem. Andreas P. Ehlers, Prof. Dr. Andreas
Supporting Information Wiley-VC 007 9 Weinheim, Germany ew ear Infrared Dyes and Fluorophores Based on Diketopyrrolopyrroles Dipl.-Chem. Georg M. Fischer, Dipl.-Chem. Andreas P. Ehlers, Prof. Dr. Andreas
«Συντήρηση αχλαδιών σε νερό. υπό την παρουσία σπόρων σιναπιού (Sinapis arvensis).»
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
SUPPORTING INFORMATION
 SUPPRTING INFRMATIN Per(-guanidino--deoxy)cyclodextrins: Synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Nikolaos Mourtzis, a Kyriaki Eliadou, a Chrysie Aggelidou,
SUPPRTING INFRMATIN Per(-guanidino--deoxy)cyclodextrins: Synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Nikolaos Mourtzis, a Kyriaki Eliadou, a Chrysie Aggelidou,
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
HOMEWORK 4 = G. In order to plot the stress versus the stretch we define a normalized stretch:
 HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Enantioselective Organocatalytic Michael Addition of Isorhodanines. to α, β-unsaturated Aldehydes
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2016 Enantioselective Organocatalytic Michael Addition of Isorhodanines to α,
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2016 Enantioselective Organocatalytic Michael Addition of Isorhodanines to α,
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ. Σπίγγος Γεώργιος
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
A Bonus-Malus System as a Markov Set-Chain. Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics
 A Bonus-Malus System as a Markov Set-Chain Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics Contents 1. Markov set-chain 2. Model of bonus-malus system 3. Example 4. Conclusions
A Bonus-Malus System as a Markov Set-Chain Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics Contents 1. Markov set-chain 2. Model of bonus-malus system 3. Example 4. Conclusions
Supplementary information:
 Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1
 Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
«Χρήσεις γης, αξίες γης και κυκλοφοριακές ρυθμίσεις στο Δήμο Χαλκιδέων. Η μεταξύ τους σχέση και εξέλιξη.»
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΑΓΡΟΝΟΜΩΝ ΚΑΙ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΓΕΩΓΡΑΦΙΑΣ ΚΑΙ ΠΕΡΙΦΕΡΕΙΑΚΟΥ ΣΧΕΔΙΑΣΜΟΥ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ: «Χρήσεις γης, αξίες γης και κυκλοφοριακές ρυθμίσεις στο Δήμο Χαλκιδέων.
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΑΓΡΟΝΟΜΩΝ ΚΑΙ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΓΕΩΓΡΑΦΙΑΣ ΚΑΙ ΠΕΡΙΦΕΡΕΙΑΚΟΥ ΣΧΕΔΙΑΣΜΟΥ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ: «Χρήσεις γης, αξίες γης και κυκλοφοριακές ρυθμίσεις στο Δήμο Χαλκιδέων.
Electronic Supplementary Information
 Electronic Supplementary Information NbCl 3 -catalyzed [2+2+2] intermolecular cycloaddition of alkynes and alkenes to 1,3-cyclohexadiene derivatives Yasushi Obora,* Keisuke Takeshita and Yasutaka Ishii*
Electronic Supplementary Information NbCl 3 -catalyzed [2+2+2] intermolecular cycloaddition of alkynes and alkenes to 1,3-cyclohexadiene derivatives Yasushi Obora,* Keisuke Takeshita and Yasutaka Ishii*
Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions
 This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Buried Markov Model Pairwise
 Buried Markov Model 1 2 2 HMM Buried Markov Model J. Bilmes Buried Markov Model Pairwise 0.6 0.6 1.3 Structuring Model for Speech Recognition using Buried Markov Model Takayuki Yamamoto, 1 Tetsuya Takiguchi
Buried Markov Model 1 2 2 HMM Buried Markov Model J. Bilmes Buried Markov Model Pairwise 0.6 0.6 1.3 Structuring Model for Speech Recognition using Buried Markov Model Takayuki Yamamoto, 1 Tetsuya Takiguchi
ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ. του φοιτητή του Τμήματος Ηλεκτρολόγων Μηχανικών και. Τεχνολογίας Υπολογιστών της Πολυτεχνικής Σχολής του. Πανεπιστημίου Πατρών
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΤΜΗΜΑ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΣΥΣΤΗΜΑΤΩΝ ΗΛΕΚΤΡΙΚΗΣ ΕΝΕΡΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΟ ΗΛΕΚΤΡΟΜΗΧΑΝΙΚΗΣ ΜΕΤΑΤΡΟΠΗΣ ΕΝΕΡΓΕΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ του φοιτητή του
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΤΜΗΜΑ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΣΥΣΤΗΜΑΤΩΝ ΗΛΕΚΤΡΙΚΗΣ ΕΝΕΡΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΟ ΗΛΕΚΤΡΟΜΗΧΑΝΙΚΗΣ ΜΕΤΑΤΡΟΠΗΣ ΕΝΕΡΓΕΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ του φοιτητή του
Electronic Supplementary Information
 Electronic Supplementary Information Unprecedented Carbon-Carbon Bond Cleavage in Nucleophilic Aziridine Ring Opening Reaction, Efficient Ring Transformation of Aziridines to Imidazolidin-4-ones Jin-Yuan
Electronic Supplementary Information Unprecedented Carbon-Carbon Bond Cleavage in Nucleophilic Aziridine Ring Opening Reaction, Efficient Ring Transformation of Aziridines to Imidazolidin-4-ones Jin-Yuan
Second Order RLC Filters
 ECEN 60 Circuits/Electronics Spring 007-0-07 P. Mathys Second Order RLC Filters RLC Lowpass Filter A passive RLC lowpass filter (LPF) circuit is shown in the following schematic. R L C v O (t) Using phasor
ECEN 60 Circuits/Electronics Spring 007-0-07 P. Mathys Second Order RLC Filters RLC Lowpass Filter A passive RLC lowpass filter (LPF) circuit is shown in the following schematic. R L C v O (t) Using phasor
Conductivity Logging for Thermal Spring Well
 /.,**. 25 +,1- **-- 0/2,,,1- **-- 0/2, +,, +/., +0 /,* Conductivity Logging for Thermal Spring Well Koji SATO +, Tadashi TAKAYA,, Tadashi CHIBA, + Nihon Chika Kenkyuusho Co. Ltd., 0/2,, Hongo, Funabashi,
/.,**. 25 +,1- **-- 0/2,,,1- **-- 0/2, +,, +/., +0 /,* Conductivity Logging for Thermal Spring Well Koji SATO +, Tadashi TAKAYA,, Tadashi CHIBA, + Nihon Chika Kenkyuusho Co. Ltd., 0/2,, Hongo, Funabashi,
Supplementary Materials for. Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides
 Supplementary Materials for Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides Indrek Kalvet, a Karl J. Bonney, a and Franziska Schoenebeck a * a Institute of Organic
Supplementary Materials for Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides Indrek Kalvet, a Karl J. Bonney, a and Franziska Schoenebeck a * a Institute of Organic
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supporting Information. Experimental section
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Emulsifying Properties of Egg Yolk as a Function of Diacylglycerol Oil
 354 Nippon Shokuhin Kagaku Kogaku Kaishi Vol. /-, No.0, -/.-0* (,**0) 38 * * Emulsifying Properties of Egg Yolk as a Function of Diacylglycerol Oil Tomoko Kawakami, Kyoko Ohashi* and Atsuko Shimada Graduate
354 Nippon Shokuhin Kagaku Kogaku Kaishi Vol. /-, No.0, -/.-0* (,**0) 38 * * Emulsifying Properties of Egg Yolk as a Function of Diacylglycerol Oil Tomoko Kawakami, Kyoko Ohashi* and Atsuko Shimada Graduate
Σύστημα επεξεργασίας, ανάλυσης και ταξινόμησης εικόνων δισδιάστατης ηλεκτροφόρησης με τεχνικές αναγνώρισης προτύπων
 ΕΘΝΙΚΟ ΚΑΙ ΚΑΠΟΔΙΣΤΡΙΑΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΤΗΛΕΠΙΚΟΙΝΩΝΙΩΝ ΔΙΑΤΜΗΜΑΤΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ "ΤΕΧΝΟΛΟΓΙΕΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΣΤΗΝ ΙΑΤΡΙΚΗ ΚΑΙ ΤΗ ΒΙΟΛΟΓΙΑ"
ΕΘΝΙΚΟ ΚΑΙ ΚΑΠΟΔΙΣΤΡΙΑΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΤΗΛΕΠΙΚΟΙΝΩΝΙΩΝ ΔΙΑΤΜΗΜΑΤΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ "ΤΕΧΝΟΛΟΓΙΕΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΣΤΗΝ ΙΑΤΡΙΚΗ ΚΑΙ ΤΗ ΒΙΟΛΟΓΙΑ"
Optimizing Microwave-assisted Extraction Process for Paprika Red Pigments Using Response Surface Methodology
 2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΓΗΑΣΜΖΜΑΣΗΚΟ ΠΡΟΓΡΑΜΜΑ ΜΔΣΑΠΣΤΥΗΑΚΩΝ ΠΟΤΓΩΝ «ΤΣΖΜΑΣΑ ΔΠΔΞΔΡΓΑΗΑ ΖΜΑΣΩΝ ΚΑΗ ΔΠΗΚΟΗΝΩΝΗΩΝ» ΣΜΖΜΑ ΜΖΥΑΝΗΚΩΝ Ζ/Τ ΚΑΗ ΠΛΖΡΟΦΟΡΗΚΖ
 ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΓΗΑΣΜΖΜΑΣΗΚΟ ΠΡΟΓΡΑΜΜΑ ΜΔΣΑΠΣΤΥΗΑΚΩΝ ΠΟΤΓΩΝ «ΤΣΖΜΑΣΑ ΔΠΔΞΔΡΓΑΗΑ ΖΜΑΣΩΝ ΚΑΗ ΔΠΗΚΟΗΝΩΝΗΩΝ» ΣΜΖΜΑ ΜΖΥΑΝΗΚΩΝ Ζ/Τ ΚΑΗ ΠΛΖΡΟΦΟΡΗΚΖ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΜΖΜΑ
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΓΗΑΣΜΖΜΑΣΗΚΟ ΠΡΟΓΡΑΜΜΑ ΜΔΣΑΠΣΤΥΗΑΚΩΝ ΠΟΤΓΩΝ «ΤΣΖΜΑΣΑ ΔΠΔΞΔΡΓΑΗΑ ΖΜΑΣΩΝ ΚΑΗ ΔΠΗΚΟΗΝΩΝΗΩΝ» ΣΜΖΜΑ ΜΖΥΑΝΗΚΩΝ Ζ/Τ ΚΑΗ ΠΛΖΡΟΦΟΡΗΚΖ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΜΖΜΑ
Πρόσκληση υποβολής προσφοράς
 Αθήνα, 5 Μαρτίου 2014 Πρόσκληση υποβολής προσφοράς από φυσικά ή νομικά πρόσωπα για την προμήθεια υλικών στην Πράξη «ΘΑΛΗΣ Εθνικό Ίδρυμα Ερευνών Βιοσύνθεση και Γενετική Επιλογή Κυκλικών Πεπτιδίων με Εν
Αθήνα, 5 Μαρτίου 2014 Πρόσκληση υποβολής προσφοράς από φυσικά ή νομικά πρόσωπα για την προμήθεια υλικών στην Πράξη «ΘΑΛΗΣ Εθνικό Ίδρυμα Ερευνών Βιοσύνθεση και Γενετική Επιλογή Κυκλικών Πεπτιδίων με Εν
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
Supporting Information
 Supporting Information Lewis acid catalyzed ring-opening reactions of methylenecyclopropanes with diphenylphosphine oxide in the presence of sulfur or selenium Min Shi,* Min Jiang and Le-Ping Liu State
Supporting Information Lewis acid catalyzed ring-opening reactions of methylenecyclopropanes with diphenylphosphine oxide in the presence of sulfur or selenium Min Shi,* Min Jiang and Le-Ping Liu State
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
SUPPLEMENTARY INFORMATION
 Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
5.4 The Poisson Distribution.
 The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
ΠΣΤΥΙΑΚΗ ΔΡΓΑΙΑ. Μειέηε Υξόλνπ Απνζηείξσζεο Κνλζέξβαο κε Τπνινγηζηηθή Ρεπζηνδπλακηθή. Αζαλαζηάδνπ Βαξβάξα
 ΣΔΥΝΟΛΟΓΙΚΟ ΔΚΠΑΙΓΔΤΣΙΚΟ ΙΓΡΤΜΑ ΘΔΑΛΟΝΙΚΗ ΥΟΛΗ ΣΔΥΝΟΛΟΓΙΑ ΣΡΟΦΙΜΩΝ & ΓΙΑΣΡΟΦΗ ΣΜΗΜΑ ΣΔΥΝΟΛΟΓΙΑ ΣΡΟΦΙΜΩΝ ΠΣΤΥΙΑΚΗ ΔΡΓΑΙΑ Μειέηε Υξόλνπ Απνζηείξσζεο Κνλζέξβαο κε Τπνινγηζηηθή Ρεπζηνδπλακηθή Αζαλαζηάδνπ Βαξβάξα
ΣΔΥΝΟΛΟΓΙΚΟ ΔΚΠΑΙΓΔΤΣΙΚΟ ΙΓΡΤΜΑ ΘΔΑΛΟΝΙΚΗ ΥΟΛΗ ΣΔΥΝΟΛΟΓΙΑ ΣΡΟΦΙΜΩΝ & ΓΙΑΣΡΟΦΗ ΣΜΗΜΑ ΣΔΥΝΟΛΟΓΙΑ ΣΡΟΦΙΜΩΝ ΠΣΤΥΙΑΚΗ ΔΡΓΑΙΑ Μειέηε Υξόλνπ Απνζηείξσζεο Κνλζέξβαο κε Τπνινγηζηηθή Ρεπζηνδπλακηθή Αζαλαζηάδνπ Βαξβάξα
Bayesian statistics. DS GA 1002 Probability and Statistics for Data Science.
 Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Longitudinal Changes in Component Processes of Working Memory
 This Accepted Manuscript has not been copyedited and formatted. The final version may differ from this version. Research Article: New Research Cognition and Behavior Longitudinal Changes in Component Processes
This Accepted Manuscript has not been copyedited and formatted. The final version may differ from this version. Research Article: New Research Cognition and Behavior Longitudinal Changes in Component Processes
Approximation of distance between locations on earth given by latitude and longitude
 Approximation of distance between locations on earth given by latitude and longitude Jan Behrens 2012-12-31 In this paper we shall provide a method to approximate distances between two points on earth
Approximation of distance between locations on earth given by latitude and longitude Jan Behrens 2012-12-31 In this paper we shall provide a method to approximate distances between two points on earth
Electrolyzed-Reduced Water as Artificial Hot Spring Water
 /-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
/-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΕΠΙΔΡΑΣΗ ΥΠΕΡΗΧΩΝ ΣΤΗΝ LISTERIA GRAYI ΣΤΟ ΓΑΛΑ: ΕΠΙΒΙΩΣΗ ΚΑΙ ΦΥΣΙΚΟΧΗΜΙΚΕΣ ΕΠΙΔΡΑΣΕΙΣ. Άρτεμις
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΕΠΙΔΡΑΣΗ ΥΠΕΡΗΧΩΝ ΣΤΗΝ LISTERIA GRAYI ΣΤΟ ΓΑΛΑ: ΕΠΙΒΙΩΣΗ ΚΑΙ ΦΥΣΙΚΟΧΗΜΙΚΕΣ ΕΠΙΔΡΑΣΕΙΣ. Άρτεμις
Chapter 1. Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes
 Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
ΓΕΩΜΕΣΡΙΚΗ ΣΕΚΜΗΡΙΩΗ ΣΟΤ ΙΕΡΟΤ ΝΑΟΤ ΣΟΤ ΣΙΜΙΟΤ ΣΑΤΡΟΤ ΣΟ ΠΕΛΕΝΔΡΙ ΣΗ ΚΤΠΡΟΤ ΜΕ ΕΦΑΡΜΟΓΗ ΑΤΣΟΜΑΣΟΠΟΙΗΜΕΝΟΤ ΤΣΗΜΑΣΟ ΨΗΦΙΑΚΗ ΦΩΣΟΓΡΑΜΜΕΣΡΙΑ
 ΕΘΝΙΚΟ ΜΕΣΟΒΙΟ ΠΟΛΤΣΕΧΝΕΙΟ ΣΜΗΜΑ ΑΓΡΟΝΟΜΩΝ-ΣΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΣΟΜΕΑ ΣΟΠΟΓΡΑΦΙΑ ΕΡΓΑΣΗΡΙΟ ΦΩΣΟΓΡΑΜΜΕΣΡΙΑ ΓΕΩΜΕΣΡΙΚΗ ΣΕΚΜΗΡΙΩΗ ΣΟΤ ΙΕΡΟΤ ΝΑΟΤ ΣΟΤ ΣΙΜΙΟΤ ΣΑΤΡΟΤ ΣΟ ΠΕΛΕΝΔΡΙ ΣΗ ΚΤΠΡΟΤ ΜΕ ΕΦΑΡΜΟΓΗ ΑΤΣΟΜΑΣΟΠΟΙΗΜΕΝΟΤ
ΕΘΝΙΚΟ ΜΕΣΟΒΙΟ ΠΟΛΤΣΕΧΝΕΙΟ ΣΜΗΜΑ ΑΓΡΟΝΟΜΩΝ-ΣΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΣΟΜΕΑ ΣΟΠΟΓΡΑΦΙΑ ΕΡΓΑΣΗΡΙΟ ΦΩΣΟΓΡΑΜΜΕΣΡΙΑ ΓΕΩΜΕΣΡΙΚΗ ΣΕΚΜΗΡΙΩΗ ΣΟΤ ΙΕΡΟΤ ΝΑΟΤ ΣΟΤ ΣΙΜΙΟΤ ΣΑΤΡΟΤ ΣΟ ΠΕΛΕΝΔΡΙ ΣΗ ΚΤΠΡΟΤ ΜΕ ΕΦΑΡΜΟΓΗ ΑΤΣΟΜΑΣΟΠΟΙΗΜΕΝΟΤ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
EE512: Error Control Coding
 EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
Statistical Inference I Locally most powerful tests
 Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Inverse trigonometric functions & General Solution of Trigonometric Equations. ------------------ ----------------------------- -----------------
 Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
ΦΩΤΟΓΡΑΜΜΕΤΡΙΚΕΣ ΚΑΙ ΤΗΛΕΠΙΣΚΟΠΙΚΕΣ ΜΕΘΟΔΟΙ ΣΤΗ ΜΕΛΕΤΗ ΘΕΜΑΤΩΝ ΔΑΣΙΚΟΥ ΠΕΡΙΒΑΛΛΟΝΤΟΣ
 AΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΕΙΔΙΚΕΥΣΗΣ ΠΡΟΣΤΑΣΙΑ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΚΑΙ ΒΙΩΣΙΜΗ ΑΝΑΠΤΥΞΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ ΦΩΤΟΓΡΑΜΜΕΤΡΙΚΕΣ
AΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΕΙΔΙΚΕΥΣΗΣ ΠΡΟΣΤΑΣΙΑ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΚΑΙ ΒΙΩΣΙΜΗ ΑΝΑΠΤΥΞΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ ΦΩΤΟΓΡΑΜΜΕΤΡΙΚΕΣ
Figure 3 Three observations (Vp, Vs and density isosurfaces) intersecting in the PLF space. Solutions exist at the two indicated points.
 φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
Μειέηε, θαηαζθεπή θαη πξνζνκνίσζε ηεο ιεηηνπξγίαο κηθξήο αλεκνγελλήηξηαο αμνληθήο ξνήο ΓΗΠΛΩΜΑΣΗΚΖ ΔΡΓΑΗΑ
 Μειέηε, θαηαζθεπή θαη πξνζνκνίσζε ηεο ιεηηνπξγίαο κηθξήο αλεκνγελλήηξηαο αμνληθήο ξνήο ΓΗΠΛΩΜΑΣΗΚΖ ΔΡΓΑΗΑ Κνηζακπφπνπινο Υ. Παλαγηψηεο Δπηβιέπσλ: Νηθφιανο Υαηδεαξγπξίνπ Καζεγεηήο Δ.Μ.Π Αζήλα, Μάξηηνο 2010
Μειέηε, θαηαζθεπή θαη πξνζνκνίσζε ηεο ιεηηνπξγίαο κηθξήο αλεκνγελλήηξηαο αμνληθήο ξνήο ΓΗΠΛΩΜΑΣΗΚΖ ΔΡΓΑΗΑ Κνηζακπφπνπινο Υ. Παλαγηψηεο Δπηβιέπσλ: Νηθφιανο Υαηδεαξγπξίνπ Καζεγεηήο Δ.Μ.Π Αζήλα, Μάξηηνο 2010
Supplementary Materials: Development of Amyloseand β-cyclodextrin-based Chiral Fluorescent Sensors Bearing Terthienyl Pendants
 S1 of S12 Supplementary Materials: Development of Amyloseand β-cyclodextrin-based Chiral Fluorescent Sensors Bearing Terthienyl Pendants Tomoyuki Ikai, Changsik Yun, Yutaka Kojima, Daisuke Suzuki, Katsuhiro
S1 of S12 Supplementary Materials: Development of Amyloseand β-cyclodextrin-based Chiral Fluorescent Sensors Bearing Terthienyl Pendants Tomoyuki Ikai, Changsik Yun, Yutaka Kojima, Daisuke Suzuki, Katsuhiro
Physical and Chemical Properties of the Nest-site Beach of the Horseshoe Crab Rehabilitated by Sand Placement
 J. Jpn. Soc. Soil Phys. No. 33, p.//0-,**/ ******* * Physical and Chemical Properties of the Nest-site Beach of the Horseshoe Crab Rehabilitated by Sand Placement Masami OHTSUBO*, Hidekazu ISHIDA**, Hisakatsu
J. Jpn. Soc. Soil Phys. No. 33, p.//0-,**/ ******* * Physical and Chemical Properties of the Nest-site Beach of the Horseshoe Crab Rehabilitated by Sand Placement Masami OHTSUBO*, Hidekazu ISHIDA**, Hisakatsu
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
The Simply Typed Lambda Calculus
 Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ
 ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ Μαυρόγιαννη Γεωργία 1, Καραντζούλα Ευαγγελία 2, Τουλιά Γεωργία 3, Κυριακόπουλος Βασίλης 4, Καδδά Όλγα 5 EΡΕΥΝΑ
ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ Μαυρόγιαννη Γεωργία 1, Καραντζούλα Ευαγγελία 2, Τουλιά Γεωργία 3, Κυριακόπουλος Βασίλης 4, Καδδά Όλγα 5 EΡΕΥΝΑ
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
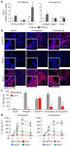 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
SUPPLEMENTAL INFORMATION. Fully Automated Total Metals and Chromium Speciation Single Platform Introduction System for ICP-MS
 Electronic Supplementary Material (ESI) for Journal of Analytical Atomic Spectrometry. This journal is The Royal Society of Chemistry 2018 SUPPLEMENTAL INFORMATION Fully Automated Total Metals and Chromium
Electronic Supplementary Material (ESI) for Journal of Analytical Atomic Spectrometry. This journal is The Royal Society of Chemistry 2018 SUPPLEMENTAL INFORMATION Fully Automated Total Metals and Chromium
Figure A.2: MPC and MPCP Age Profiles (estimating ρ, ρ = 2, φ = 0.03)..
 Supplemental Material (not for publication) Persistent vs. Permanent Income Shocks in the Buffer-Stock Model Jeppe Druedahl Thomas H. Jørgensen May, A Additional Figures and Tables Figure A.: Wealth and
Supplemental Material (not for publication) Persistent vs. Permanent Income Shocks in the Buffer-Stock Model Jeppe Druedahl Thomas H. Jørgensen May, A Additional Figures and Tables Figure A.: Wealth and
Σχέση µεταξύ της Μεθόδου των ερµατοπτυχών και της Βιοηλεκτρικής Αντίστασης στον Υπολογισµό του Ποσοστού Σωµατικού Λίπους
 Ερευνητική Aναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµότόµος 1(3), 244 251 ηµοσιεύτηκε: 2 8 εκεµβρίου 2003 Inquiries in Sport & Physical Education Volume 1(3), 244 251 Released: December 28, 2003 www.hape.gr/emag.asp
Ερευνητική Aναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµότόµος 1(3), 244 251 ηµοσιεύτηκε: 2 8 εκεµβρίου 2003 Inquiries in Sport & Physical Education Volume 1(3), 244 251 Released: December 28, 2003 www.hape.gr/emag.asp
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής
 Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Sampling Basics (1B) Young Won Lim 9/21/13
 Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks
 98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
Metal-free Oxidative Coupling of Amines with Sodium Sulfinates: A Mild Access to Sulfonamides
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting information for Metal-free Oxidative Coupling of Amines with Sodium Sulfinates:
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting information for Metal-free Oxidative Coupling of Amines with Sodium Sulfinates:
Συστήματα Διαχείρισης Βάσεων Δεδομένων
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Συστήματα Διαχείρισης Βάσεων Δεδομένων Φροντιστήριο 9: Transactions - part 1 Δημήτρης Πλεξουσάκης Τμήμα Επιστήμης Υπολογιστών Tutorial on Undo, Redo and Undo/Redo
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Συστήματα Διαχείρισης Βάσεων Δεδομένων Φροντιστήριο 9: Transactions - part 1 Δημήτρης Πλεξουσάκης Τμήμα Επιστήμης Υπολογιστών Tutorial on Undo, Redo and Undo/Redo
Statistics 104: Quantitative Methods for Economics Formula and Theorem Review
 Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Section 8.3 Trigonometric Equations
 99 Section 8. Trigonometric Equations Objective 1: Solve Equations Involving One Trigonometric Function. In this section and the next, we will exple how to solving equations involving trigonometric functions.
99 Section 8. Trigonometric Equations Objective 1: Solve Equations Involving One Trigonometric Function. In this section and the next, we will exple how to solving equations involving trigonometric functions.
ST5224: Advanced Statistical Theory II
 ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
College of Life Science, Dalian Nationalities University, Dalian , PR China.
 Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2018 Postsynthetic modification
Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2018 Postsynthetic modification
Supplementary Information
 Electronic upplementary Material (EI) for Photochemical & Photobiological ciences. This journal is The Royal ociety of Chemistry and wner ocieties 214 upplementary Information elective and sensitive fluorescence-shift
Electronic upplementary Material (EI) for Photochemical & Photobiological ciences. This journal is The Royal ociety of Chemistry and wner ocieties 214 upplementary Information elective and sensitive fluorescence-shift
Phys460.nb Solution for the t-dependent Schrodinger s equation How did we find the solution? (not required)
 Phys460.nb 81 ψ n (t) is still the (same) eigenstate of H But for tdependent H. The answer is NO. 5.5.5. Solution for the tdependent Schrodinger s equation If we assume that at time t 0, the electron starts
Phys460.nb 81 ψ n (t) is still the (same) eigenstate of H But for tdependent H. The answer is NO. 5.5.5. Solution for the tdependent Schrodinger s equation If we assume that at time t 0, the electron starts
Electronic Supplementary Information
 Electronic Supplementary Information Cucurbit[7]uril host-guest complexes of the histamine H 2 -receptor antagonist ranitidine Ruibing Wang and Donal H. Macartney* Department of Chemistry, Queen s University,
Electronic Supplementary Information Cucurbit[7]uril host-guest complexes of the histamine H 2 -receptor antagonist ranitidine Ruibing Wang and Donal H. Macartney* Department of Chemistry, Queen s University,
Η ΤΟΞΙΚΟΤΗΤΑ ΤΟΥ ΒΟΡΙΟΥ(B) ΣΤΗΝ ΚΑΛΛΙΕΡΓΕΙΑ ΤΗΣ ΤΟΜΑΤΑΣ
 ΑΛΕΞΑΝΔΡΕΙΟ Τ.Ε.Ι. ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΑΣ ΓΕΩΠΟΝΙΑΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΚΑΙ ΔΙΑΤΡΟΦΗΣ ΤΜΗΜΑ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΑΝΑΤΟΜΙΑΣ ΚΑΙ ΦΥΣΙΟΛΟΓΙΑΣ ΦΥΤΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η ΤΟΞΙΚΟΤΗΤΑ ΤΟΥ ΒΟΡΙΟΥ(B)
ΑΛΕΞΑΝΔΡΕΙΟ Τ.Ε.Ι. ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΑΣ ΓΕΩΠΟΝΙΑΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΚΑΙ ΔΙΑΤΡΟΦΗΣ ΤΜΗΜΑ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΑΝΑΤΟΜΙΑΣ ΚΑΙ ΦΥΣΙΟΛΟΓΙΑΣ ΦΥΤΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η ΤΟΞΙΚΟΤΗΤΑ ΤΟΥ ΒΟΡΙΟΥ(B)
EPL 603 TOPICS IN SOFTWARE ENGINEERING. Lab 5: Component Adaptation Environment (COPE)
 EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
Matrices and vectors. Matrix and vector. a 11 a 12 a 1n a 21 a 22 a 2n A = b 1 b 2. b m. R m n, b = = ( a ij. a m1 a m2 a mn. def
 Matrices and vectors Matrix and vector a 11 a 12 a 1n a 21 a 22 a 2n A = a m1 a m2 a mn def = ( a ij ) R m n, b = b 1 b 2 b m Rm Matrix and vectors in linear equations: example E 1 : x 1 + x 2 + 3x 4 =
Matrices and vectors Matrix and vector a 11 a 12 a 1n a 21 a 22 a 2n A = a m1 a m2 a mn def = ( a ij ) R m n, b = b 1 b 2 b m Rm Matrix and vectors in linear equations: example E 1 : x 1 + x 2 + 3x 4 =
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
Supporting Information One-Pot Approach to Chiral Chromenes via Enantioselective Organocatalytic Domino Oxa-Michael-Aldol Reaction
 Supporting Information ne-pot Approach to Chiral Chromenes via Enantioselective rganocatalytic Domino xa-michael-aldol Reaction Hao Li, Jian Wang, Timiyin E-Nunu, Liansuo Zu, Wei Jiang, Shaohua Wei, *
Supporting Information ne-pot Approach to Chiral Chromenes via Enantioselective rganocatalytic Domino xa-michael-aldol Reaction Hao Li, Jian Wang, Timiyin E-Nunu, Liansuo Zu, Wei Jiang, Shaohua Wei, *
ΜΔΛΔΣΖ ΔΝΓΟΣΡΑΥΤΝΖ Δ ΥΑΛΤΒΔ ΘΔΡΜΖ ΔΛΑΖ
 ΔΘΝΗΚΟ ΜΔΣΟΒΗΟ ΠΟΛΤΣΔΥΝΔΗΟ ΔΙΑΣΜΗΜΑΣΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΣΑΠΣΤΥΙΑΚΩΝ ΠΟΤΔΩΝ (Δ.Π.Μ..): "ΔΠΗΣΖΜΖ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΛΗΚΧΝ" ΜΔΛΔΣΖ ΔΝΓΟΣΡΑΥΤΝΖ Δ ΥΑΛΤΒΔ ΘΔΡΜΖ ΔΛΑΖ ΜΕΣΑΠΣΤΥΙΑΚΗ ΕΡΓΑΙΑ ΕΔΡΒΑ ΑΗΜΗΛΗΑΝΟ Διπλωματούτος
ΔΘΝΗΚΟ ΜΔΣΟΒΗΟ ΠΟΛΤΣΔΥΝΔΗΟ ΔΙΑΣΜΗΜΑΣΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΣΑΠΣΤΥΙΑΚΩΝ ΠΟΤΔΩΝ (Δ.Π.Μ..): "ΔΠΗΣΖΜΖ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΛΗΚΧΝ" ΜΔΛΔΣΖ ΔΝΓΟΣΡΑΥΤΝΖ Δ ΥΑΛΤΒΔ ΘΔΡΜΖ ΔΛΑΖ ΜΕΣΑΠΣΤΥΙΑΚΗ ΕΡΓΑΙΑ ΕΔΡΒΑ ΑΗΜΗΛΗΑΝΟ Διπλωματούτος
Daewoo Technopark A-403, Dodang-dong, Wonmi-gu, Bucheon-city, Gyeonggido, Korea LM-80 Test Report
 LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
1 h, , CaCl 2. pelamis) 58.1%, (Headspace solid -phase microextraction and gas chromatography -mass spectrometry,hs -SPME - Vol. 15 No.
 2 0 1 5 7 Journal of Chinese Institute of Food Science and Technology Vol. 15 No. 7 Jul. 2 0 1 5 1,2 1 1 1 1* ( 1 315211 2 315502) : ( ) : (E-Nose), - : GC-MS, 20.98% 2.5%, 53.15% 11.2%, 0. 51% 71.86%
2 0 1 5 7 Journal of Chinese Institute of Food Science and Technology Vol. 15 No. 7 Jul. 2 0 1 5 1,2 1 1 1 1* ( 1 315211 2 315502) : ( ) : (E-Nose), - : GC-MS, 20.98% 2.5%, 53.15% 11.2%, 0. 51% 71.86%
Correction Table for an Alcoholometer Calibrated at 20 o C
 An alcoholometer is a device that measures the concentration of ethanol in a water-ethanol mixture (often in units of %abv percent alcohol by volume). The depth to which an alcoholometer sinks in a water-ethanol
An alcoholometer is a device that measures the concentration of ethanol in a water-ethanol mixture (often in units of %abv percent alcohol by volume). The depth to which an alcoholometer sinks in a water-ethanol
Jesse Maassen and Mark Lundstrom Purdue University November 25, 2013
 Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ
 ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ Γηπισκαηηθή Δξγαζία ηνπ Φνηηεηή ηνπ ηκήκαηνο Ζιεθηξνιόγσλ Μεραληθώλ θαη Σερλνινγίαο Ζιεθηξνληθώλ
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ Γηπισκαηηθή Δξγαζία ηνπ Φνηηεηή ηνπ ηκήκαηνο Ζιεθηξνιόγσλ Μεραληθώλ θαη Σερλνινγίαο Ζιεθηξνληθώλ
Pyrrolo[2,3-d:5,4-d']bisthiazoles: Alternate Synthetic Routes and a Comparative Study to Analogous Fused-ring Bithiophenes
![Pyrrolo[2,3-d:5,4-d']bisthiazoles: Alternate Synthetic Routes and a Comparative Study to Analogous Fused-ring Bithiophenes Pyrrolo[2,3-d:5,4-d']bisthiazoles: Alternate Synthetic Routes and a Comparative Study to Analogous Fused-ring Bithiophenes](/thumbs/92/108867714.jpg) SUPPORTING INFORMATION Pyrrolo[2,3-d:5,4-d']bisthiazoles: Alternate Synthetic Routes and a Comparative Study to Analogous Fused-ring Bithiophenes Eric J. Uzelac, Casey B. McCausland, and Seth C. Rasmussen*
SUPPORTING INFORMATION Pyrrolo[2,3-d:5,4-d']bisthiazoles: Alternate Synthetic Routes and a Comparative Study to Analogous Fused-ring Bithiophenes Eric J. Uzelac, Casey B. McCausland, and Seth C. Rasmussen*
9-amino-(9-deoxy)cinchona alkaloids-derived novel chiral phase-transfer catalysts
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2014 9-amino-(9-deoxy)cinchona alkaloids-derived novel chiral phase-transfer
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2014 9-amino-(9-deoxy)cinchona alkaloids-derived novel chiral phase-transfer
the total number of electrons passing through the lamp.
 1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
