Molecular Structure: matching and kinematics
|
|
|
- Χρυσάνθη Δράκος
- 6 χρόνια πριν
- Προβολές:
Transcript
1 Molecular Structure: matching and kinematics Ioannis Z. Emiris Dept. of Informatics & Telecoms, University of Athens Algs in Struc.BioInfo 16
2 Outline 03. Chemical bonds 06. Structure types, aminoacids, Ramachandran plot 19. Structure comparison 27. Databases, and prediction 33. Kinematics and Rigid transforms (39. Parameterization) 50. Motion planning: Configuration space 59. Degrees of freedom 64. Appendix: Energy, Ramachandran, CASP 70. Structure matching, geometric hashing 77. Protein folding 85. Docking Reading: Wikipedia for RMSD: Root-mean-square deviation. Choset, Kavraki et al. Principles of Robot Motion, Chap. 3, and E.
3 Chemical bonds
4 Chemical Bonds Covalent: Share electrons. Polar, e.g. HO, HN, HCl. Non-polar, e.g. H 2. Simple (allows rotation), or multiple (double, triple etc). Non-covalent or Ionic (or salt): transfer electrons. E.g. N acl Hydrogen: between covalently bonded H to electronegative atom, and electronegative atom, eg. O, N. Similar to non-covalent: O H O = Van der Waals: push-away A if r 3.5Å: disallows atoms to occupy same space (strong push when nearby).
5 Breakup Energy [kcal / mole] bond in void in H 2 O covalent non-covalent 90 3 H-bond Van der Waals
6 Structure types
7 Structure types Primary: ATCCGTG, FQRRTVQILQT Secondary: α-helix, β-sheet Super-secondary: α-hairpin, β-hairpin, β α β Tertiary: (3.9Å, 2.2Å, -45.1Å),.... Overall fold. E.g. N, H α, C, C β form regular tetrahedron centered at C α. Mirror symmetries isomers (proteins w/one isomer always). Quaternary: several monomers (domains) van der Waals.
8 Primary/tertiary structure of the 20 aminoacids. Backbone skeleton is: top-c, center (C α ), N-at-left. min: Glycine, special: Proline.
9 Properties of the 20 aminoacids Nonpolar not-h 2 O soluble (hydrophobic) Polar are: globally neutral, acidic(-), or basic(+). Charged are: acidic (e-acceptor) or basic (e-donor), not always polar.
10 α-helix DNA: usually B-DNA in cells, radius 10Å; Z-, A-DNA less frequent, less standard helix. Proteins: Usually right-handed spiral (αr); Sidechains usually lie inside the helix Glycine may form left-handed helix αl. Each C α advances by: 100 o, 1.5Å. i.e. 3.6 residues per turn of the helix. Rigidity by H-bonds: (C=O) i (HN) i+4, and inward hydrophobic, outward hydrophilic faces.
11 β-sheet Parallel xor antiparallel (twisted). Composed of 2 (almost) coplanar strands. φ, ψ angles differ by π. Each C α advances by: 3.5Å Rigidity due to H-bonds: C=O HN, of neighboring strands
12 Antiparallel vs Parallel
13 (β-sheet variants) β-barrel: Sheet-end has right-handed twist, folds on itself. Figure: human retinol-binding protein (1RBP): 8 anti strands form barrel, binding vitamin A. If residue with 2 H-bonds bulge
14 (Rare helices) residues per turn, bind i + 4 thus narrower, longer usually at end of α-helix Π 4 residues per turn, bind i + 5 squat and constrained
15 Protein tertiary structure N, H α, C, C β form Tetrahedron, center C α. Rotation around single bonds N-C α (φ) and C α -C (ψ) one exception: proline (only ψ). angle ω around peptide bond has 2 states: trans: ω 180 o is usual cis: ω 0 o is rare, mostly at proline
16 Average rigid elements Bond Angle Mean [ o ] N-C α C o C α C-N o C-N-C α o C α N-H o Bond Length N-C α C α C C-N N-H Mean [Å] 1.46 Å 1.53 Å 1.33 Å 0.98 Å
17 Sasisekharan-Ramakrishnan-Ramachandran diagram Describes allowed mainchain conformations. Horizontal φ, vertical ψ, typically ω = 180 o. parallel β P, twisted β T ; right-handed α, left-handed L, 3 10, Π helices. Exception: Gly (no limitation), Pro (side chain back to backbone).
18 Ramachandran diagram: example structure types: α, β, Gly for protein 2ACY [Lesk].
19 Structure comparison
20 Measure difference of matched sets Hypotheses: pointsets of equal cardinality, given correspondance (match) Definition. (coordinate) Root Mean Square Deviation (c-rmsd) RMSD = 1 n n i=1 x i y i 2, where x i, y i R 3 are (C α ) atom coordinates in SAME coordinate frame. Lemma. c-rmsd satisfies the triangular inequality. Hence it defines a distance metric.
21 Optimal Alignment of matched sets Problem. Find translation and rotation minimizing c-rmsd. 1. Translate to common origin by subtracting from x i s centroid x c = 1 n n i=1 x i, x i R 3, and subtracting y c from all y i s; overall = O(n). 2. Rotate to optimal alignment by 3 3 rotation matrix Q : Q T Q = I. Also should have det Q = 1. Deterministic linear algebra (SVD) algorithm [Kabsch]: O(n). Lemma: optimal translation can be decoupled from rotation optimization. Proof: for any Q, optimal translation brings center of mass to origin.
22 Matrix algebra Let X = [x 1,..., x n ] T, Y = [y 1,..., y n ] T R n 3, then RMSD(X, Y ) = 1 n X Y F, where M 2 F = i,j M 2 ij = tr(m T M), is the Frobenius norm, tr(a) = i A ii is the trace of matrix A = [A ij ]. Recall rotated vector is v T Q or Qv, for column vector v R 3.
23 Optimal rotation Assume common centroid = 0, X, Y R n 3 : RMSD(X, Y ) = min Q Y XQ F, Q T Q = I, Q = 1. We apply: tr(a + B) =tr(a)+tr(b), tr(a) =tr(a T ), (AB) T = B T A T, Lemma A: tr(a T A) = A 2 F = AT 2 F = tr(aat ), and Lemma T: tr(ab) = ij A ij B ji = tr(ba), for A, B of equal size. Proposition. Optimizing rotation Q R 3 3 reduces to max Q tr(q T X T Y ). Proof: Y XQ 2 F = tr[(y XQ)T (Y XQ)] = = tr(y T Y ) + tr(x T X) 2tr(Q T X T Y ), where tr[(xq) T (XQ)] = tr(x T X) by Lemma A, and tr[(y ) T (XQ)] = tr[(xq) T Y ].
24 Singular Value Decomposition Recall SVD: X T Y = UΣV T, U T U = V T V = I, Σ = σ σ σ 3 where : σ 1 σ 2 σ 3, U, V, Σ are 3 3 like X T Y, and singular values σ i = e i 0, e i are eigenvalues(x T Y ). We wish to find Q that maximizes: tr(q T X T Y ) = tr(q T UΣ V T ) = tr(v T Q T UΣ) tr(σ). 2nd equality by Lem. T; inequality since M = V T Q T U is orthonormal M ij 1 tr(mσ) = i M ii σ i i σ i. Thm. Maximum occurs at M = I Q = UV T. If det Q = 1 then Q reflection, hence negate Q 33 to get rotation. Overall complexity = O(n).
25 Algorithm Input: pointsets X, Y R n 3 of n corresponding points. Output: minimum RMSD of translated and rotated sets. Algorithm. x c n i=1 x i /n, y c n i=1 y i /n. X {x x c : x X}, Y {y y c : y Y }. SVD: X T Y = UΣV T. Optional: Check σ 3 > 0, where Σ = diag[σ 1, σ 2, σ 3 ]. Q U V T. If det Q < 0 then Q [U 1, U 2, U 3 ] V T. // U i : ith column Return X Q Y F / n // or ni=1 Qx i y i 2 /n
26 distance-rmsd Assume that k distances d i, i = 1,..., k are known between point-pairs in X and between the corresponding pairs in Y, denoted d i, i = 1,..., k. Defn. For k matched distances, there is a distance-rmsd 2 = 1 k k i=1 Drawback: Computed in O(k) = O(n 2 ). (d i d i )2, k ( n 2 ). Lem. d-rmsd invariant under rigid transforms: translate, rotate, reflect. d-rmsd is a metric in (Euclidean) R r space; but then one point represents a conformation and its mirror image. Please check [Guibas?]: c-rmsd / n d-rmsd 2 c-rmsd.
27 Databases, and prediction
28 Databases Protein Data Bank (PDB) ( Structure information and retrieval File starts with protein name, author, maybe secondary structure Omits H-atoms Example: Hemoglobin, residue of Argynine: ATOM N ARG ATOM CA ARG ATOM C ARG ATOM O ARG ATOM CB ARG Protein fold classification into hierarchies: SCOP (Structural Classification of Proteins), cf next slide [Murzin et al 95, Andreeva et al 04] CATH (domains) (Class, architecture, topology, homology) [Orengo et al 97, Pearl et al 05] FSSP (DALI offers structural alignment) [Holm,Sander 96] CE (structural alignment)
29 SCOP Hierarchy Lowest level: individual protein domains (from PDB) families of homologues: similar structure, sequence, (function) imply common evolutionary origin superfamilies: families of similar structure and function, weak evolutionary relationship folds: superfamilies with common folding topology Highest level: classes: α, β, α + β, α/β (α and β) and small proteins Homology of structures expresses common ancestry: either evolutionary: evolved from structure in common ancestor (wings of bats and arms of primates), or developmental: from same tissue in embryonal development (ovaries of female and testicles of male humans).
30 SCOP example 1 Root SCOP 2 Class α/β, mainly parallel β-sheets (β α β units) 3 Fold Flavodoxin-like: 3 layers, α/β/α; parallel β-sheet of 5 strands, order Superfamily Flavoproteins 5 Family Flavodoxin-related binds FMN 6 Protein Flavodoxin 7 Species Clostridium beijerinckii [Lesk,p.224]
31 SCOP size In July 2001, SCOP contained 13,220 PDB entries, in 31,474 domains: Class families superfamilies folds All-α proteins All-β proteins α/β proteins α + β proteins Multi-domain membrane, cell-surface Small proteins Total
32 Rigid-body kinematics: Motivation
33 Molecular kinematics Given a rigid body with specific degrees of freedom (e.g. dihedral angles about covalent bonds), its kinematics describe the allowed motions under certain geometric constraints (distances, angles etc) Modeling of constraints as an algebraic / optimization problem. Applications: structure determination of small (sub)molecules, dimension-reduction during docking, pharmacophore matching. There s many small molecules: most (about 15%) with 4 dof, < 10% with > 10 dof, out of 730,000 w/rotational dof [Irwin-Shoichet 04]
34 Rigid transforms
35 Rigid (Euclidean) transformations Preserve distances and angles. Translation d R 3, x x + d. Rotation R SO(3) : R 1 = R T, det R = 1, x Rx. R 1 : rotation by negative angle. R 1 by θ 1, R 2 by θ 2 R 1 R 2 by θ 1 + θ 2. Reflection R : det R = 1 (reflection in R 2 takes body out of the plane) Scaling and Shearing are NOT rigid.
36 2D transforms Rotation, scaling, shearing: [ ] [ cos θ sin θ sx 0, sin θ cos θ 0 s y ] (typically s x, s y > 0), [ 1 a 0 1 ]. T = cos θ sin θ 0 sin θ cos θ d : homogeneous transform: translation by d, rotation (by θ) : R SO(2), R 1 SO(2), R 1 = R T, det R = 1. cos θ sin θ 0 sin θ cos θ d x y 1 i+1 = x y 1 i
37 Classification Linear transforms represented by matrix-vector multiplication: Rotation, reflection, scaling, shearing (NOT translation). They preserve linear combinations of points. Affine transform M = L + T where L is linear and T is translation. Def. 4 basic affine transforms: rotation, scaling, shearing, translation Thm. Every affine transformation written as a combination/sequence of these 4 basic affine transformations. Cor. Reflection is affine combination of 4 basics.
38 3D motion Rotation is 3 3 matrix parameterized by only 3 free elements (e.g. Euler angles in next slide, or Quaternions), not 9. Transformation between frames: cos θ sin θ cos α sin θ sin α a cos θ sin θ cos θ cos α cos θ sin α a sin θ 0 sin α cos α r Coordinate frame X i Y i Z i associated to i-th rigid link, joint allows motion between links i, i + 1. x y z 1 i+1 = x y z 1 i 4 Denavit-Hartenberg parameters (see below for details): α angle between axes Z i, Z i+1, θ about joint i (dihedral), link i of length a measured between the Z-axes, offset r at joint i 1 measured along Z i.
39 (Parameterization)
40 Rotation Matrix with Euler Angles Rotation about z0 by α + Rotation about y1 by β + Rotation about x2 by γ T 0 3 = T 0 1 T 1 2 T 2 3 = cos(α) sin(α) 0 cos(β) 0 sin(β) = sin(α) cos(α) cos(γ) sin(γ) = sin(β) 0 cos(β) 0 sin(γ) cos(γ) cos(α) cos(β) cos(α) sin(β) sin(γ) sin(α) cos(γ) cos(α) sin(β) cos(γ) + sin(α) sin(γ) sin(α) cos(β) sin(α) sin(β) sin(γ) + cos(α) cos(γ) sin(α) sin(β) cos(γ) cos(α) sin(γ) sin(β) cos(β) sin(γ) cos(β) cos(γ) [Choset,Kavraki et al. Principles of Robot Motion, Chap.E]
41 Denavit-Hartenberg Method (1955)
42 Denavit-Hartenberg Matrix For transforming two coordinate frames: Rot(Z i, θ i ) + Trans(Z i, r i ) + Trans(X i+1, a i ) + Rot(X i+1, α i ) A i = A i = cos(θ i ) sin(θ i ) 0 0 sin(θ i ) cos(θ i ) r i a i 0 cos(α i ) sin(α i ) 0 0 sin(α i ) cos(α i ) cos(θ i ) sin(θ i ) cos(α i ) sin(θ i ) sin(α i ) a i cos(θ i ) sin(θ i ) cos(θ i ) cos(α i ) cos(θ i ) sin(α i ) a i sin(θ i ) 0 sin(α i ) cos(α i ) r i X i -axis is normal to Z i and Z i 1 axes, at distance = a i 1.
43 Kinematic Chains Two types of rigid mechanisms / robots / linkages / manipulators: Serial linkage of rigid bodies connected by movable joints. Open chain, i.e. no loops. Wish to analyze motion of last link (end effector) in terms of other links. Apply Matrix multiplication: T all = T 1 T 2 T n. Parallel robots: all linkages connected to same end-effector. T 1 = = T n, where T i depends on parameters. Kinematics Problems: T i contains unknown (e.g. angle θ i ). Inverse kinematics: Given T all, compute the θ i. Trivial for parallel robots, hard for serial robots. Forward/direct kinematics: Given the T i, compute T all. Trivial for serial manipulators, hard for parallel manipulators.
44 Planar 2-R Manipulator
45 Planar 2R: parameters D-H Parameters: (stars denote variables) Link a i α i r i θ i 1 a θ 1 2 a θ 2 A 1 = Homogeneous Transformation c 1 s 1 0 a 1 c 1 s 1 c 1 0 a 1 s , A 2 = c 2 s 2 0 a 2 c 2 s 2 c 2 0 a 2 s T 0 2 = A 1A 2 = T 0 1 = A 1 c 12 s 12 0 a 1 c 1 + a 2 c 12 s 12 c 12 0 a 1 s 1 + a 2 s c 12, s 12 refer to θ 1 + θ 2.,
46 Planar 2R: kinematics Direct Kinematics x 0 = a 1 cos(θ 1 ) + a 2 cos(θ 1 + θ 2 ) y 0 = a 1 sin(θ 1 ) + a 2 sin(θ 1 + θ 2 ) θ 1 = cos 1 Inverse Kinematics y x 2 + y 2 cos 1 x2 + y 2 + a 2 1 a2 2 2a 1 x 2 + y 2 θ 2 = cos 1 x2 + y 2 a 2 1 a2 2 2a 1 a 2
47 3-Link RPP Manipulator
48 RPP: parameters D-H Parameters: (stars denote variables) Link a i α i r i θ i d 1 θ o d d 3 0 A 1 = Homogeneous Transformation c 1 s s 1 c d 1, A 2 = d 2, A 3 = d T 0 3 = A 1A 2 A 3 = c 1 0 s 1 s 1 d 3 s 1 0 c 1 c 1 d d 1 + d
49 RPP: kinematics Direct Kinematics: x 0 = sin(θ 1 )d 3 y 0 = cos(θ 1 )d 3 z 0 = d 1 + d 2 Inverse Kinematics: θ 1 = tan 1 ( x y ) d 2 = z d 1 d 3 = x 2 + y 2
50 Motion planning
51 Εισαγωγή Ερωτήματα σχετικά με τον σχεδιασμό κίνησης (motion planning) ενός ρομποτικού μηχανισμού: Πόση πληροφορία χρειάζεται για να προσδιοριστεί η θέση κάθε σημείου του ρομπότ; Πώς θα αναπαρασταθεί η παραπάνω πληροφορία; Ποιες είναι οι μαθηματικές ιδιότητες της αναπαράστασης της πληροφορίας; Πώς θα λάβουμε υπ όψιν τα εμπόδια στον σχεδιασμό των κινήσεων; [Choset, Kavraki et al. Principles of Robot Motion, Chapter 3]
52 Βασικές έννοιες Διαμόρφωση (robot configuration, molecule conformation): πλήρης προσδιορισμός της θέσης (π.χ. 3 συντεταγμένες) κάθε σημείου του ρομπότ. Χώρος διαμορφώσεων (Configuration space, C-space): Ο χώρος όλων των πιθανών διαμορφώσεων του ρομπότ, όπου καθε διαμόρφωση αντιστοιχεί σε ένα σημείο του χώρου. Βαθμοί ελευθερίας (Degrees of freedom): Ο αριθμός των παραμέτρων που απαιτούνται για να προσδιοριστεί μία διαμόρφωση. Ισοδύναμα, η διάσταση του χώρου διαμορφώσεων. Χώρος εργασίας (Workspace): Ο φυσικός χώρος που είναι προσβάσιμος από το ρομπότ, τυπικά 3Δ. Προσοχή: Χώρος εργασίας Χώρος διαμορφώσεων.
53 Παράδειγμα 1: Ρομπότ-δίσκος Ρομπότ-δίσκος, δεδομένης ακτίνας r, το οποίο κινείται στο δισδιάστατο επίπεδο R 2. Διαμόρφωση: q = (x, y) αρκεί να προσδιοριστεί το κέντρο του ρομπότ, άρα C-space R 2. Για κάθε διαμόρφωση μπορούμε να υπολογίσουμε τα σημεία που καταλαμβάνει το ρομπότ ως εξής: R(x, y) = {(x, y ) R 2 (x x ) 2 + (y y ) 2 r 2 }, r = ακτίνα του ρομπότ. Μπορούμε να ορίσουμε τον χώρο διαμορφώσεων και τον χώρο εργασίας. Είναι και οι δύο υποσύνολα του R 2, αλλά είναι διαφορετικοί!
54 Παράδειγμα 2: Βραχίονας με δύο αρθρώσεις
55 Παράδειγμα 2: Βραχίονας με δύο αρθρώσεις Διαμόρφωση: η θέση του χεριού (elbow up / down δηλ. θ 2 ) δεν αρκεί: χρειάζονται οι γωνίες και των 2 αρθρώσεων: q = (θ 1, θ 2 ). Κάθε άρθρωση μπορεί να περιστραφεί σε ένα μοναδιαίο κύκλο S 1 χώρος διαμορφώσεων Q = S 1 S 1 = T 2 δηλ. δισδιάστατος τόρος. χώρος εργασίας = ένας δίσκος R 2 (εικόνα δεξιά).
56 Εμπόδια Εμποδια χώρου διαμορφώσεων (C-space obstacles): Διαμορφώσεις q όπου το ρομπότ R(q) συγκρούεται με εμπόδιο W i : O i = {q Q R(q) W i }. Ελευθερος χώρος διαμορφώσεων (free C-space): Q free = Q \ ( i O i ) Ελεύθερο μονοπάτι (free path): Μονοπάτι χωρίς συγκρούσεις με εμπόδια που δεν περιλαμβάνει ούτε τα ακραία σημεία του Q free. Δίνεται από παραμετροποίηση: c : [0, 1] Q free. Ημι-ελεύθερο μονοπάτι (semifree path): Οπως το ελεύθερο, αλλά μπορεί να περιλαβει ακραία σημεία (όριο) του Q free : c : [0, 1] Closure(Q free ).
57 Παράδειγμα 1 (με εμπόδια) (1) Κυκλικό ρομπότ και πολυγωνικό εμπόδιο στο R 2. (2) το ρομπότ διατρέχει το εμπόδιο του χώρου εργασίας (workspace obstacle). Ελέγχουμε συγκεκριμένα σημεία. (3) Η τροχιά του κέντρου ορίζει το εμπόδιο στον χώρο διαμορφώσεων (C-space obstacle), όπου το ρομπότ = σημείο. Επαυξημένο πολύγωνο = άθροισμα Minkowski του αρχικού + δίσκο
58 Παράδειγμα 2 (με εμπόδια) A A Για τα εμπόδια στον χώρο διαμορφώσεων, θεωρούμε σύνολο διαμορφώσεων και για καθεμία υπολογίζουμε αν προκαλεί σύγκρουση. Ο βραχίονας έχει 2 αρθρώσεις: θ 1 = 0 στον άξονα x, θ 2 = 0 στον x, αμφότερες CCW. One point is fixed (center of left fig.). [Choset,Kavraki et al. Sec.3.2.2]
59 Degrees of freedom (dof) Holonomic constraints are expressed purely as a function of configuration variables (and possibly time); e.g. distances: f(q, t) = 0. For each holonomic constraint, degrees of freedom are reduced by 1: n coordinates and m holonomic constraints lead to n m dof. Non-Holonomic constraints imply not all dof are controllable, e.g. cars with 2 controls but 3 dof.
60 Planar rigid body Robot translates and rotates. A, B, C: 3 distinct points fixed to the body of robot. 6 coordinates : (x A, y A ), (x B, y B ), (x C, y C ) 3 holonomic constraints (distances): d(a, B) = (x A x B ) 2 + (y A y B ) 2 d(a, C) = (x A x C ) 2 + (y A y C ) 2 d(b, C) = (x B x C ) 2 + (y B y C ) 2 Thus, 3 dof q = (x A, y A, θ) (x A, y A ) : position of point A θ : orientation of robot Configuration space: Q = R 2 S 1
61 Open-chain jointed robot (serial mechanism) Usually, add the dof s at each joint Common joints with 1 degree of freedom revolute (R): rotating about an axis prismatic (P): translating along an axis Common joint with 3 dof s spherical (ball-and-socket)
62 Closed-chain jointed robot (parallel mechanism) 1 stationary (base) + (k 1) movable = k links System starts with N(k 1) dof (before accounting for joints): Each movable link has N dof s N = 6 for spatial mechanism N = 3 for planar mechanism Each joint places N f i constraints f i : dof s at joint i f i = 1 for prismatic/revolute joint Grübler s (or Kuzbach) formula for mobility (dof), assuming all constraints are independent: M = N(k 1) n i=1 (N f i ) = N(k n 1) + n i=1 f i.
63 Planar mechanism with 6 links: Examples B E F C A D N = 3 dof; k = 6 links A,..., F ; n = 7 revolute joints, each f i = 1. Mobility by Grübler/Kuzbach: M = 3(6 7 1) + 7 = 1 Stewart platform: 6 legs, each 5 links, 6 R/P joints. Hence k = links, n = 36 joints. Mobility by Grübler/Kuzbach M = (6 1) = 6.
64 Appendix: Chemistry, Energy, Structure
65 Chemistry basics b X a a =atomic number= #protons = # electrons, b = atomic weight= atom weight H-atom weight #protons +#neutrons. weight of H-atom = 1 dalton := g. 1 mole = Avogadro atoms = atoms = b g. Atoms with common a and different b are isotopes: 1 H 1, 12 C 6, 14 N 7, 16 O 8, 31 P 15, 32 S 16, 39 K 19.
66 Total Energy Total potential (for Molecular Mechanics) is the sum of the following: Covalent bonds: length of bond b, where standard len 0 : c b (len b len 0 ) 2, angle between bonds, standard angle θ 0 : c a (θ a θ 0 ) 2, dihedral angle φ i, standard φ 0 : c i (1 + cos(φ i φ 0 )). Non-bond interaction (i, j) ( Lennard-Jones (mainly van der Waals): D ij ( c ij r ) 12 ( c ij ij electrostatic: r ij = distance, q i = charge: q i q j e r e 0 r ij. r ij ) 6 ), Constants: c b 500 kcal/mole, c a 5 kcal/mole, Dielectric Constant = e r e 0.
67 Ramachandran diagram (stats) 20-residue average except Gly / Pro
68 (Tertiary) structure determination Modeling geometry flexibility (required by function) n backbone residues (say 100), k 3 conformations each = molecules However nature finds favorable conformation, fast (Levinthal s paradox)
69 CASP benchmark Critical Assessment of Structure Prediction Organizes blind test of protein structure, given sequence. Runs on a two year cycle. Experimental results are kept secret until predictions are submitted. secondary structure: helix H, (extended) strand E, other 3D structure predicted by assembling predicted helices, strands of sheet [Janin]
70 Structure matching
71 Rigid Matching Finding best transform ie. yielding max/bio-favorable superposition. Dependent on sequence-order: Matching set [Taylor-Orengo 89] (Dynamic Programming SSAP). fragments [Vriend-Sander] follow sequence order. FSSP-DALI [Holm,Sander 93], CE [Bourne,Shindyalov 98] Independent of Sequence (unlabeled points, different cardinalities) Geometric hashing (from vision): finds translation, rotation, scaling maxclique in SSE graph (by 2ary elements) [Mitchel et al] [Koch et al] Sequence independence: - 3d task vs essentially linear task. - Simultaneous match of sequence / structure is better + Finds non-sequential motifs eg. binding sites + works with partial / disconnected input
72 Geometric Hashing: 2D preprocess Preprocess each pointset (model) in database: pair (points #4, #1 below), define a reference frame: Compute coordinates (x, y) of all points in this frame, store [model, frame] in entry Hash(x, y). Storing 3 hash entries (2 shown by arrows) in 2D
73 Geometric Hashing: 2D query Online processing of query pointset (image): I. Pick reference frame (defined by 2 points): compute coordinates of all query points in this frame. II. Hash query points: for every data point in its hash-entry, cast a vote for the corresponding [model, frame] III [model, transform] with high scores induce potential match: optimize transform by least-squares (or RMSD on matched points) Hashed points vote for each [model,frame] pair in their hash entries (2 arrows shown)
74 Geometric Hashing: Complexity Parameters. M = #structures in database (models), n = #points per structure/model, c = 1 + #points to define a frame: c = 3 in 2D, c = 4 in 3D. Time complexity. preprocess = O(Mn c ), online query = O(Hn c ), where H = #complexity of checking one hashtable entry. H = O(1) typically when Space = O(Mn c ), good hashing; or can be H = O(Mn c ) for small/unlucky tables. [eclass/eggrafa/apallaktikh/wolfson-rigoutsos 99]
75 Geometric hashing: generalization Idea: Given two objects each with n unlabeled points: Each Pair of almost-congruent triangles defines 3D rigid transform (congruent/similar: invariant under translation, rotation, scaling) For each candidate transform, count superposed points. For best candidates, find RMSD on matched pairs, keep the best. Complexity O(n 7 ) (if we exploit backbone geometry: n 3 ) [Wolfson slides] Against database: 0. point (residue), define local neighborhood. 1. Geometric Hashing gives seed matches. 2. Cluster seed matches by merging matched points 3. Compare RMSDs of clusters; extend better clusters until solution Extra: store features into [model, frame, features]
76 Flexible Alignment Motivation. Mutations/docking imply conformational change Hinge and shear motion of domains [Lesk] Existing work 3D curve matching [Schwartz,Sharir 87], using splines [Wolfson et al 91] Dock [Leach,Kutz]. FlexX (dock), FlexS (structures) use anchors [Lengauer,Lemmen,Klebe 98] small-molecule database search [Rigoutsos,Platt,Califano 96] Pose clustering [Verbitsky,Wolfson,Nussinov 99]. Known hinges, hashing [Fligelman,Nussinov,Wolfson 00] FlexProt [Shatsky,Nussinov,Wolfson 02]
77 Optional further topics
78 Protein folding
79 Folding Protein folding: process by which a polypeptide folds from its mrna unfolded state into its native state, i.e. characteristic (and functional) 3D structure. Result determined by amino acid sequence. This is Anfinsen s, or 2nd, dogma [Anfinsen 73]
80 Folding factors Folding depends on solvent (water or lipid bilayer) concentration of salts temperature presence of molecular chaperones [Lee,Tsai 05] Folding is affected by external fields (electric, magnetic) molecular crowding, limitation of space.
81 Disruption of the native state Proteins denaturate: thermally unstable: high/low temperatures solutes, extremes of ph, mechanical forces, chemical denaturants Denatured protein: random coil, no secondary or tertiary structure. Chaperones: protect from denaturation; help to fold. Incorrect folding and aggregated proteins cause: prion-related illnesses amyloid-related illnesses familial amyloid cardiomyopathy or polyneuropathy intracytoplasmic aggregation diseases proteopathy diseases Therapy: protein replacement, pharmaceutical chaperones
82 Folding speed Levinthal s paradox [Levinthal 68]: Folding is NP-hard. fast! (through a series of intermediate states) Yet, proteins fold proline isomerization: minutes or hours small single-domain proteins: milliseconds or microseconds
83 Computational methods Energy landscape: principle of minimal frustration describes protein folding by leveling the free-energy landscape high-dimensional phase-space where manifolds take several topological forms [Robson,Vaithilingham 08] supported by computational simulation of model proteins, and experimental studies [Bryngelson,Onuchic,Socci,Wolynes 95]
84 Modeling of protein folding Molecular Dynamics (MD): studying protein folding and dynamics in silico. First: implicit solvent model and Umbrella Sampling. High computational cost: with explicit water peptides and very small proteins. larger proteins only dynamics of the structure or high-temperature unfolding long-time folding processes approximations or simplifications in protein models (pseudo-atoms representing groups of atoms) Distributed computing projects: project.
85 Docking
86 Definitions Receptor: receiving molecule, usually protein or other biopolymer. Ligand: complementary partner molecule, binds to receptor; most often small molecules but could be another biopolymer. Docking: simulation of candidate ligand binding to receptor. Binding mode: orientation of ligand relative to receptor and conformation of ligand and receptor when bound together. Pose: a candidate binding mode. Scoring: process of evaluating particular pose by counting number of favorable intermolecular interactions e.g. hydrogen bonds, hydrophobic contacts. Ranking: classifying which ligands are most likely to interact favorably to particular receptor based on predicted free-energy of binding.
87 Significance of Docking Predicting: strength of association (binding affinity) between two molecules; strength and type (e.g., agonism vs antagonism) of signal; binding orientation of small molecule drug candidates to their protein targets. Docking: computationally simulate the molecular recognition process. optimized conformation and relative orientation for protein and ligand s.t. the free energy is minimized.
88 Docking approaches Shape complementarity: matching technique that describes the protein and the ligand as complementary surfaces. [Goldman,Wipke 00], [Meng,Shoichet,Kuntz 04], [Morris,Goodsell,Halliday,Huey,Hart,Belew,Olson 98] Simulation: simulate the actual docking process in which the ligand-protein pairwise interaction energies are calculated [Feig,Onufriev,Lee,Im,Case,Brooks 04]
89 Shape complementarity Describe that makes protein and ligand dockable [Shoichet,Kuntz,Bodian 04]: Molecular surface/ complementary surface: receptor - solvent-accessible surface area, ligand - matching surface Hydrophobic features of the protein: turns in the main-chain atoms. Fourier shape descriptor technique [Cai,Shao,Maigret 02], [Morris,Najmanovich,Kahraman,Thornton 05], [Kahraman,Morris,Laskowski,Thornton 07] Advantages: fast and robust; scalable to protein-protein interactions; amenable to pharmacophore based approaches. Disadvantage: cannot accurately model the movements or dynamic changes.
90 Simulation Protein and ligand are separated by some physical distance, ligand finds its position into the protein s active site after a certain number of moves. After each total energy of the system is calculated. Moves: translations rotations internal changes to the ligand s structure (incl. torsion angle rotations) Advantages: easy to incorporate ligand flexibility; process physically closer to reality. Disadvantage: slow
91 Mechanics of docking Determine structure of the protein by X-ray crystallography NMR spectroscopy Docking program: search algorithm scoring function
92 Search algorithm Search space: all possible orientations and conformations of protein paired with ligand. enumerate all possible distortions of each molecule, all possible rotational and translational orientations of the ligand relative to the protein. In practice: flexible ligand (most programs) flexible protein receptor Search Strategies: systematic or stochastic torsional searches about rotatable bonds molecular dynamics simulations genetic algorithms to evolve new low energy conformations
93 Ligand flexibility Conformations of the ligand may be generated without receptor, then docked [Kearsley,Underwood,Sheridan,Miller 94] on-the-fly in the presence of the receptor binding cavity [Friesner et al 04] with full rotational flexibility of every dihedral angle (fragment based docking) [Zsoldos,Reid,Simon,Sadjad,Johnson 07] Select low energy conformations: force field energy evaluation (usual) [Wang,Pang 07] knowledge-based methods [Klebe,Mietzner 94]
94 Receptor flexibility large number of degrees of freedom [Cerqueira,Bras,Fernandes,Ramos 09] Emulate receptor flexibility: Multiple static structures for the same protein in different conformations [Totrov,Abagyan 08] Rotamer libraries of amino acid side chains that surround the binding cavity [Hartmann,Antes,Lengauer 09], [Taylor,Jewsbury,Essex 03]
95 Scoring function Input: pose ( snapshot of ligand + protein) Output: number (likelihood of binding) Scoring functions analyze: energy of the pose (physics-based molecular mechanics force fields): a low (negative) energy likely binding interaction potential fit of the pose (databases of protein-ligand complexes)
96 Using Databases X-ray crystallography: Many: proteins + high affinity ligands Less: proteins + low affinity ligands false positive hits Solution: recalculate the energy of the top scoring poses (Generalized Born, Poisson-Boltzmann [Feig,Onufriev,Lee,Im,Case,Brooks 04])
97 Applications Hit identification: docking combined with scoring function used to quickly screen large databases of potential drugs to identify molecules that are likely to bind to protein target of interest (virtual screening). Lead optimization: predict where and in which relative orientation a ligand binds to a protein (binding mode or pose). Bio-remediation: predict pollutants that can be degraded by enzymes [Suresh,Kumar,Kumar,Singh 08].
Molecular Structure: matching and kinematics
 Molecular Structure: matching and kinematics Ioannis Z. Emiris Dept. of Informatics & Telecoms, University of Athens Algs in Struc.BioInfo 17 Outline 03. Structure types, aminoacids, Ramachandran plot
Molecular Structure: matching and kinematics Ioannis Z. Emiris Dept. of Informatics & Telecoms, University of Athens Algs in Struc.BioInfo 17 Outline 03. Structure types, aminoacids, Ramachandran plot
Reminders: linear functions
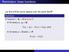 Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
3.4 SUM AND DIFFERENCE FORMULAS. NOTE: cos(α+β) cos α + cos β cos(α-β) cos α -cos β
 3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Approximation of distance between locations on earth given by latitude and longitude
 Approximation of distance between locations on earth given by latitude and longitude Jan Behrens 2012-12-31 In this paper we shall provide a method to approximate distances between two points on earth
Approximation of distance between locations on earth given by latitude and longitude Jan Behrens 2012-12-31 In this paper we shall provide a method to approximate distances between two points on earth
HOMEWORK 4 = G. In order to plot the stress versus the stretch we define a normalized stretch:
 HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Section 8.3 Trigonometric Equations
 99 Section 8. Trigonometric Equations Objective 1: Solve Equations Involving One Trigonometric Function. In this section and the next, we will exple how to solving equations involving trigonometric functions.
99 Section 8. Trigonometric Equations Objective 1: Solve Equations Involving One Trigonometric Function. In this section and the next, we will exple how to solving equations involving trigonometric functions.
Phys460.nb Solution for the t-dependent Schrodinger s equation How did we find the solution? (not required)
 Phys460.nb 81 ψ n (t) is still the (same) eigenstate of H But for tdependent H. The answer is NO. 5.5.5. Solution for the tdependent Schrodinger s equation If we assume that at time t 0, the electron starts
Phys460.nb 81 ψ n (t) is still the (same) eigenstate of H But for tdependent H. The answer is NO. 5.5.5. Solution for the tdependent Schrodinger s equation If we assume that at time t 0, the electron starts
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3
 Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Numerical Analysis FMN011
 Numerical Analysis FMN011 Carmen Arévalo Lund University carmen@maths.lth.se Lecture 12 Periodic data A function g has period P if g(x + P ) = g(x) Model: Trigonometric polynomial of order M T M (x) =
Numerical Analysis FMN011 Carmen Arévalo Lund University carmen@maths.lth.se Lecture 12 Periodic data A function g has period P if g(x + P ) = g(x) Model: Trigonometric polynomial of order M T M (x) =
Nowhere-zero flows Let be a digraph, Abelian group. A Γ-circulation in is a mapping : such that, where, and : tail in X, head in
 Nowhere-zero flows Let be a digraph, Abelian group. A Γ-circulation in is a mapping : such that, where, and : tail in X, head in : tail in X, head in A nowhere-zero Γ-flow is a Γ-circulation such that
Nowhere-zero flows Let be a digraph, Abelian group. A Γ-circulation in is a mapping : such that, where, and : tail in X, head in : tail in X, head in A nowhere-zero Γ-flow is a Γ-circulation such that
2 Composition. Invertible Mappings
 Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
EE512: Error Control Coding
 EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
derivation of the Laplacian from rectangular to spherical coordinates
 derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
b. Use the parametrization from (a) to compute the area of S a as S a ds. Be sure to substitute for ds!
 MTH U341 urface Integrals, tokes theorem, the divergence theorem To be turned in Wed., Dec. 1. 1. Let be the sphere of radius a, x 2 + y 2 + z 2 a 2. a. Use spherical coordinates (with ρ a) to parametrize.
MTH U341 urface Integrals, tokes theorem, the divergence theorem To be turned in Wed., Dec. 1. 1. Let be the sphere of radius a, x 2 + y 2 + z 2 a 2. a. Use spherical coordinates (with ρ a) to parametrize.
Areas and Lengths in Polar Coordinates
 Kiryl Tsishchanka Areas and Lengths in Polar Coordinates In this section we develop the formula for the area of a region whose boundary is given by a polar equation. We need to use the formula for the
Kiryl Tsishchanka Areas and Lengths in Polar Coordinates In this section we develop the formula for the area of a region whose boundary is given by a polar equation. We need to use the formula for the
9.09. # 1. Area inside the oval limaçon r = cos θ. To graph, start with θ = 0 so r = 6. Compute dr
 9.9 #. Area inside the oval limaçon r = + cos. To graph, start with = so r =. Compute d = sin. Interesting points are where d vanishes, or at =,,, etc. For these values of we compute r:,,, and the values
9.9 #. Area inside the oval limaçon r = + cos. To graph, start with = so r =. Compute d = sin. Interesting points are where d vanishes, or at =,,, etc. For these values of we compute r:,,, and the values
Section 9.2 Polar Equations and Graphs
 180 Section 9. Polar Equations and Graphs In this section, we will be graphing polar equations on a polar grid. In the first few examples, we will write the polar equation in rectangular form to help identify
180 Section 9. Polar Equations and Graphs In this section, we will be graphing polar equations on a polar grid. In the first few examples, we will write the polar equation in rectangular form to help identify
6.1. Dirac Equation. Hamiltonian. Dirac Eq.
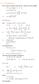 6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
( ) 2 and compare to M.
 Problems and Solutions for Section 4.2 4.9 through 4.33) 4.9 Calculate the square root of the matrix 3!0 M!0 8 Hint: Let M / 2 a!b ; calculate M / 2!b c ) 2 and compare to M. Solution: Given: 3!0 M!0 8
Problems and Solutions for Section 4.2 4.9 through 4.33) 4.9 Calculate the square root of the matrix 3!0 M!0 8 Hint: Let M / 2 a!b ; calculate M / 2!b c ) 2 and compare to M. Solution: Given: 3!0 M!0 8
The Simply Typed Lambda Calculus
 Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Practice Exam 2. Conceptual Questions. 1. State a Basic identity and then verify it. (a) Identity: Solution: One identity is csc(θ) = 1
 Conceptual Questions. State a Basic identity and then verify it. a) Identity: Solution: One identity is cscθ) = sinθ) Practice Exam b) Verification: Solution: Given the point of intersection x, y) of the
Conceptual Questions. State a Basic identity and then verify it. a) Identity: Solution: One identity is cscθ) = sinθ) Practice Exam b) Verification: Solution: Given the point of intersection x, y) of the
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 6/5/2006
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα είναι μικρότεροι το 1000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Διάρκεια: 3,5 ώρες Καλή
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα είναι μικρότεροι το 1000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Διάρκεια: 3,5 ώρες Καλή
[1] P Q. Fig. 3.1
![[1] P Q. Fig. 3.1 [1] P Q. Fig. 3.1](/thumbs/79/80362156.jpg) 1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
Μονοβάθμια Συστήματα: Εξίσωση Κίνησης, Διατύπωση του Προβλήματος και Μέθοδοι Επίλυσης. Απόστολος Σ. Παπαγεωργίου
 Μονοβάθμια Συστήματα: Εξίσωση Κίνησης, Διατύπωση του Προβλήματος και Μέθοδοι Επίλυσης VISCOUSLY DAMPED 1-DOF SYSTEM Μονοβάθμια Συστήματα με Ιξώδη Απόσβεση Equation of Motion (Εξίσωση Κίνησης): Complete
Μονοβάθμια Συστήματα: Εξίσωση Κίνησης, Διατύπωση του Προβλήματος και Μέθοδοι Επίλυσης VISCOUSLY DAMPED 1-DOF SYSTEM Μονοβάθμια Συστήματα με Ιξώδη Απόσβεση Equation of Motion (Εξίσωση Κίνησης): Complete
Areas and Lengths in Polar Coordinates
 Kiryl Tsishchanka Areas and Lengths in Polar Coordinates In this section we develop the formula for the area of a region whose boundary is given by a polar equation. We need to use the formula for the
Kiryl Tsishchanka Areas and Lengths in Polar Coordinates In this section we develop the formula for the area of a region whose boundary is given by a polar equation. We need to use the formula for the
6.3 Forecasting ARMA processes
 122 CHAPTER 6. ARMA MODELS 6.3 Forecasting ARMA processes The purpose of forecasting is to predict future values of a TS based on the data collected to the present. In this section we will discuss a linear
122 CHAPTER 6. ARMA MODELS 6.3 Forecasting ARMA processes The purpose of forecasting is to predict future values of a TS based on the data collected to the present. In this section we will discuss a linear
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
CRASH COURSE IN PRECALCULUS
 CRASH COURSE IN PRECALCULUS Shiah-Sen Wang The graphs are prepared by Chien-Lun Lai Based on : Precalculus: Mathematics for Calculus by J. Stuwart, L. Redin & S. Watson, 6th edition, 01, Brooks/Cole Chapter
CRASH COURSE IN PRECALCULUS Shiah-Sen Wang The graphs are prepared by Chien-Lun Lai Based on : Precalculus: Mathematics for Calculus by J. Stuwart, L. Redin & S. Watson, 6th edition, 01, Brooks/Cole Chapter
SCHOOL OF MATHEMATICAL SCIENCES G11LMA Linear Mathematics Examination Solutions
 SCHOOL OF MATHEMATICAL SCIENCES GLMA Linear Mathematics 00- Examination Solutions. (a) i. ( + 5i)( i) = (6 + 5) + (5 )i = + i. Real part is, imaginary part is. (b) ii. + 5i i ( + 5i)( + i) = ( i)( + i)
SCHOOL OF MATHEMATICAL SCIENCES GLMA Linear Mathematics 00- Examination Solutions. (a) i. ( + 5i)( i) = (6 + 5) + (5 )i = + i. Real part is, imaginary part is. (b) ii. + 5i i ( + 5i)( + i) = ( i)( + i)
Section 7.6 Double and Half Angle Formulas
 09 Section 7. Double and Half Angle Fmulas To derive the double-angles fmulas, we will use the sum of two angles fmulas that we developed in the last section. We will let α θ and β θ: cos(θ) cos(θ + θ)
09 Section 7. Double and Half Angle Fmulas To derive the double-angles fmulas, we will use the sum of two angles fmulas that we developed in the last section. We will let α θ and β θ: cos(θ) cos(θ + θ)
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
On a four-dimensional hyperbolic manifold with finite volume
 BULETINUL ACADEMIEI DE ŞTIINŢE A REPUBLICII MOLDOVA. MATEMATICA Numbers 2(72) 3(73), 2013, Pages 80 89 ISSN 1024 7696 On a four-dimensional hyperbolic manifold with finite volume I.S.Gutsul Abstract. In
BULETINUL ACADEMIEI DE ŞTIINŢE A REPUBLICII MOLDOVA. MATEMATICA Numbers 2(72) 3(73), 2013, Pages 80 89 ISSN 1024 7696 On a four-dimensional hyperbolic manifold with finite volume I.S.Gutsul Abstract. In
Example Sheet 3 Solutions
 Example Sheet 3 Solutions. i Regular Sturm-Liouville. ii Singular Sturm-Liouville mixed boundary conditions. iii Not Sturm-Liouville ODE is not in Sturm-Liouville form. iv Regular Sturm-Liouville note
Example Sheet 3 Solutions. i Regular Sturm-Liouville. ii Singular Sturm-Liouville mixed boundary conditions. iii Not Sturm-Liouville ODE is not in Sturm-Liouville form. iv Regular Sturm-Liouville note
The challenges of non-stable predicates
 The challenges of non-stable predicates Consider a non-stable predicate Φ encoding, say, a safety property. We want to determine whether Φ holds for our program. The challenges of non-stable predicates
The challenges of non-stable predicates Consider a non-stable predicate Φ encoding, say, a safety property. We want to determine whether Φ holds for our program. The challenges of non-stable predicates
PARTIAL NOTES for 6.1 Trigonometric Identities
 PARTIAL NOTES for 6.1 Trigonometric Identities tanθ = sinθ cosθ cotθ = cosθ sinθ BASIC IDENTITIES cscθ = 1 sinθ secθ = 1 cosθ cotθ = 1 tanθ PYTHAGOREAN IDENTITIES sin θ + cos θ =1 tan θ +1= sec θ 1 + cot
PARTIAL NOTES for 6.1 Trigonometric Identities tanθ = sinθ cosθ cotθ = cosθ sinθ BASIC IDENTITIES cscθ = 1 sinθ secθ = 1 cosθ cotθ = 1 tanθ PYTHAGOREAN IDENTITIES sin θ + cos θ =1 tan θ +1= sec θ 1 + cot
Inverse trigonometric functions & General Solution of Trigonometric Equations. ------------------ ----------------------------- -----------------
 Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Parametrized Surfaces
 Parametrized Surfaces Recall from our unit on vector-valued functions at the beginning of the semester that an R 3 -valued function c(t) in one parameter is a mapping of the form c : I R 3 where I is some
Parametrized Surfaces Recall from our unit on vector-valued functions at the beginning of the semester that an R 3 -valued function c(t) in one parameter is a mapping of the form c : I R 3 where I is some
Εισαγωγή στις πρωτεΐνες Δομή πρωτεϊνών Ταξινόμηση βάσει δομής Βάσεις με δομές πρωτεϊνών Ευθυγράμμιση δομών Πρόβλεψη 2D δομής Πρόβλεψη 3D δομής
 Εισαγωγή στις πρωτεΐνες Δομή πρωτεϊνών Ταξινόμηση βάσει δομής Βάσεις με δομές πρωτεϊνών Ευθυγράμμιση δομών Πρόβλεψη 2D δομής Πρόβλεψη 3D δομής Τι είναι η πρωτεΐνη Τι εννοούμε με δομή πρωτεϊνών Οικογένειες
Εισαγωγή στις πρωτεΐνες Δομή πρωτεϊνών Ταξινόμηση βάσει δομής Βάσεις με δομές πρωτεϊνών Ευθυγράμμιση δομών Πρόβλεψη 2D δομής Πρόβλεψη 3D δομής Τι είναι η πρωτεΐνη Τι εννοούμε με δομή πρωτεϊνών Οικογένειες
Written Examination. Antennas and Propagation (AA ) April 26, 2017.
 Written Examination Antennas and Propagation (AA. 6-7) April 6, 7. Problem ( points) Let us consider a wire antenna as in Fig. characterized by a z-oriented linear filamentary current I(z) = I cos(kz)ẑ
Written Examination Antennas and Propagation (AA. 6-7) April 6, 7. Problem ( points) Let us consider a wire antenna as in Fig. characterized by a z-oriented linear filamentary current I(z) = I cos(kz)ẑ
TMA4115 Matematikk 3
 TMA4115 Matematikk 3 Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet Trondheim Spring 2010 Lecture 12: Mathematics Marvellous Matrices Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet
TMA4115 Matematikk 3 Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet Trondheim Spring 2010 Lecture 12: Mathematics Marvellous Matrices Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Lecture 2. Soundness and completeness of propositional logic
 Lecture 2 Soundness and completeness of propositional logic February 9, 2004 1 Overview Review of natural deduction. Soundness and completeness. Semantics of propositional formulas. Soundness proof. Completeness
Lecture 2 Soundness and completeness of propositional logic February 9, 2004 1 Overview Review of natural deduction. Soundness and completeness. Semantics of propositional formulas. Soundness proof. Completeness
Capacitors - Capacitance, Charge and Potential Difference
 Capacitors - Capacitance, Charge and Potential Difference Capacitors store electric charge. This ability to store electric charge is known as capacitance. A simple capacitor consists of 2 parallel metal
Capacitors - Capacitance, Charge and Potential Difference Capacitors store electric charge. This ability to store electric charge is known as capacitance. A simple capacitor consists of 2 parallel metal
ΚΥΠΡΙΑΚΟΣ ΣΥΝΔΕΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY 21 ος ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ Δεύτερος Γύρος - 30 Μαρτίου 2011
 Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Strain gauge and rosettes
 Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS
 CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
Solutions to Exercise Sheet 5
 Solutions to Eercise Sheet 5 jacques@ucsd.edu. Let X and Y be random variables with joint pdf f(, y) = 3y( + y) where and y. Determine each of the following probabilities. Solutions. a. P (X ). b. P (X
Solutions to Eercise Sheet 5 jacques@ucsd.edu. Let X and Y be random variables with joint pdf f(, y) = 3y( + y) where and y. Determine each of the following probabilities. Solutions. a. P (X ). b. P (X
Second Order Partial Differential Equations
 Chapter 7 Second Order Partial Differential Equations 7.1 Introduction A second order linear PDE in two independent variables (x, y Ω can be written as A(x, y u x + B(x, y u xy + C(x, y u u u + D(x, y
Chapter 7 Second Order Partial Differential Equations 7.1 Introduction A second order linear PDE in two independent variables (x, y Ω can be written as A(x, y u x + B(x, y u xy + C(x, y u u u + D(x, y
Chapter 7 Transformations of Stress and Strain
 Chapter 7 Transformations of Stress and Strain INTRODUCTION Transformation of Plane Stress Mohr s Circle for Plane Stress Application of Mohr s Circle to 3D Analsis 90 60 60 0 0 50 90 Introduction 7-1
Chapter 7 Transformations of Stress and Strain INTRODUCTION Transformation of Plane Stress Mohr s Circle for Plane Stress Application of Mohr s Circle to 3D Analsis 90 60 60 0 0 50 90 Introduction 7-1
Fractional Colorings and Zykov Products of graphs
 Fractional Colorings and Zykov Products of graphs Who? Nichole Schimanski When? July 27, 2011 Graphs A graph, G, consists of a vertex set, V (G), and an edge set, E(G). V (G) is any finite set E(G) is
Fractional Colorings and Zykov Products of graphs Who? Nichole Schimanski When? July 27, 2011 Graphs A graph, G, consists of a vertex set, V (G), and an edge set, E(G). V (G) is any finite set E(G) is
Block Ciphers Modes. Ramki Thurimella
 Block Ciphers Modes Ramki Thurimella Only Encryption I.e. messages could be modified Should not assume that nonsensical messages do no harm Always must be combined with authentication 2 Padding Must be
Block Ciphers Modes Ramki Thurimella Only Encryption I.e. messages could be modified Should not assume that nonsensical messages do no harm Always must be combined with authentication 2 Padding Must be
ST5224: Advanced Statistical Theory II
 ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
DESIGN OF MACHINERY SOLUTION MANUAL h in h 4 0.
 DESIGN OF MACHINERY SOLUTION MANUAL -7-1! PROBLEM -7 Statement: Design a double-dwell cam to move a follower from to 25 6, dwell for 12, fall 25 and dwell for the remader The total cycle must take 4 sec
DESIGN OF MACHINERY SOLUTION MANUAL -7-1! PROBLEM -7 Statement: Design a double-dwell cam to move a follower from to 25 6, dwell for 12, fall 25 and dwell for the remader The total cycle must take 4 sec
Chapter 6: Systems of Linear Differential. be continuous functions on the interval
 Chapter 6: Systems of Linear Differential Equations Let a (t), a 2 (t),..., a nn (t), b (t), b 2 (t),..., b n (t) be continuous functions on the interval I. The system of n first-order differential equations
Chapter 6: Systems of Linear Differential Equations Let a (t), a 2 (t),..., a nn (t), b (t), b 2 (t),..., b n (t) be continuous functions on the interval I. The system of n first-order differential equations
Statistical Inference I Locally most powerful tests
 Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
the total number of electrons passing through the lamp.
 1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
Homework 3 Solutions
 Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
EPL 603 TOPICS IN SOFTWARE ENGINEERING. Lab 5: Component Adaptation Environment (COPE)
 EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
Trigonometric Formula Sheet
 Trigonometric Formula Sheet Definition of the Trig Functions Right Triangle Definition Assume that: 0 < θ < or 0 < θ < 90 Unit Circle Definition Assume θ can be any angle. y x, y hypotenuse opposite θ
Trigonometric Formula Sheet Definition of the Trig Functions Right Triangle Definition Assume that: 0 < θ < or 0 < θ < 90 Unit Circle Definition Assume θ can be any angle. y x, y hypotenuse opposite θ
Partial Differential Equations in Biology The boundary element method. March 26, 2013
 The boundary element method March 26, 203 Introduction and notation The problem: u = f in D R d u = ϕ in Γ D u n = g on Γ N, where D = Γ D Γ N, Γ D Γ N = (possibly, Γ D = [Neumann problem] or Γ N = [Dirichlet
The boundary element method March 26, 203 Introduction and notation The problem: u = f in D R d u = ϕ in Γ D u n = g on Γ N, where D = Γ D Γ N, Γ D Γ N = (possibly, Γ D = [Neumann problem] or Γ N = [Dirichlet
ω ω ω ω ω ω+2 ω ω+2 + ω ω ω ω+2 + ω ω+1 ω ω+2 2 ω ω ω ω ω ω ω ω+1 ω ω2 ω ω2 + ω ω ω2 + ω ω ω ω2 + ω ω+1 ω ω2 + ω ω+1 + ω ω ω ω2 + ω
 0 1 2 3 4 5 6 ω ω + 1 ω + 2 ω + 3 ω + 4 ω2 ω2 + 1 ω2 + 2 ω2 + 3 ω3 ω3 + 1 ω3 + 2 ω4 ω4 + 1 ω5 ω 2 ω 2 + 1 ω 2 + 2 ω 2 + ω ω 2 + ω + 1 ω 2 + ω2 ω 2 2 ω 2 2 + 1 ω 2 2 + ω ω 2 3 ω 3 ω 3 + 1 ω 3 + ω ω 3 +
0 1 2 3 4 5 6 ω ω + 1 ω + 2 ω + 3 ω + 4 ω2 ω2 + 1 ω2 + 2 ω2 + 3 ω3 ω3 + 1 ω3 + 2 ω4 ω4 + 1 ω5 ω 2 ω 2 + 1 ω 2 + 2 ω 2 + ω ω 2 + ω + 1 ω 2 + ω2 ω 2 2 ω 2 2 + 1 ω 2 2 + ω ω 2 3 ω 3 ω 3 + 1 ω 3 + ω ω 3 +
Lecture 15 - Root System Axiomatics
 Lecture 15 - Root System Axiomatics Nov 1, 01 In this lecture we examine root systems from an axiomatic point of view. 1 Reflections If v R n, then it determines a hyperplane, denoted P v, through the
Lecture 15 - Root System Axiomatics Nov 1, 01 In this lecture we examine root systems from an axiomatic point of view. 1 Reflections If v R n, then it determines a hyperplane, denoted P v, through the
5. Choice under Uncertainty
 5. Choice under Uncertainty Daisuke Oyama Microeconomics I May 23, 2018 Formulations von Neumann-Morgenstern (1944/1947) X: Set of prizes Π: Set of probability distributions on X : Preference relation
5. Choice under Uncertainty Daisuke Oyama Microeconomics I May 23, 2018 Formulations von Neumann-Morgenstern (1944/1947) X: Set of prizes Π: Set of probability distributions on X : Preference relation
10/3/ revolution = 360 = 2 π radians = = x. 2π = x = 360 = : Measures of Angles and Rotations
 //.: Measures of Angles and Rotations I. Vocabulary A A. Angle the union of two rays with a common endpoint B. BA and BC C. B is the vertex. B C D. You can think of BA as the rotation of (clockwise) with
//.: Measures of Angles and Rotations I. Vocabulary A A. Angle the union of two rays with a common endpoint B. BA and BC C. B is the vertex. B C D. You can think of BA as the rotation of (clockwise) with
Problem Set 9 Solutions. θ + 1. θ 2 + cotθ ( ) sinθ e iφ is an eigenfunction of the ˆ L 2 operator. / θ 2. φ 2. sin 2 θ φ 2. ( ) = e iφ. = e iφ cosθ.
 Chemistry 362 Dr Jean M Standard Problem Set 9 Solutions The ˆ L 2 operator is defined as Verify that the angular wavefunction Y θ,φ) Also verify that the eigenvalue is given by 2! 2 & L ˆ 2! 2 2 θ 2 +
Chemistry 362 Dr Jean M Standard Problem Set 9 Solutions The ˆ L 2 operator is defined as Verify that the angular wavefunction Y θ,φ) Also verify that the eigenvalue is given by 2! 2 & L ˆ 2! 2 2 θ 2 +
Homework 8 Model Solution Section
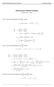 MATH 004 Homework Solution Homework 8 Model Solution Section 14.5 14.6. 14.5. Use the Chain Rule to find dz where z cosx + 4y), x 5t 4, y 1 t. dz dx + dy y sinx + 4y)0t + 4) sinx + 4y) 1t ) 0t + 4t ) sinx
MATH 004 Homework Solution Homework 8 Model Solution Section 14.5 14.6. 14.5. Use the Chain Rule to find dz where z cosx + 4y), x 5t 4, y 1 t. dz dx + dy y sinx + 4y)0t + 4) sinx + 4y) 1t ) 0t + 4t ) sinx
5.4 The Poisson Distribution.
 The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
Απόκριση σε Μοναδιαία Ωστική Δύναμη (Unit Impulse) Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο. Απόστολος Σ.
 Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο The time integral of a force is referred to as impulse, is determined by and is obtained from: Newton s 2 nd Law of motion states that the action
Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο The time integral of a force is referred to as impulse, is determined by and is obtained from: Newton s 2 nd Law of motion states that the action
Jesse Maassen and Mark Lundstrom Purdue University November 25, 2013
 Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
Ordinal Arithmetic: Addition, Multiplication, Exponentiation and Limit
 Ordinal Arithmetic: Addition, Multiplication, Exponentiation and Limit Ting Zhang Stanford May 11, 2001 Stanford, 5/11/2001 1 Outline Ordinal Classification Ordinal Addition Ordinal Multiplication Ordinal
Ordinal Arithmetic: Addition, Multiplication, Exponentiation and Limit Ting Zhang Stanford May 11, 2001 Stanford, 5/11/2001 1 Outline Ordinal Classification Ordinal Addition Ordinal Multiplication Ordinal
Abstract Storage Devices
 Abstract Storage Devices Robert König Ueli Maurer Stefano Tessaro SOFSEM 2009 January 27, 2009 Outline 1. Motivation: Storage Devices 2. Abstract Storage Devices (ASD s) 3. Reducibility 4. Factoring ASD
Abstract Storage Devices Robert König Ueli Maurer Stefano Tessaro SOFSEM 2009 January 27, 2009 Outline 1. Motivation: Storage Devices 2. Abstract Storage Devices (ASD s) 3. Reducibility 4. Factoring ASD
4.6 Autoregressive Moving Average Model ARMA(1,1)
 84 CHAPTER 4. STATIONARY TS MODELS 4.6 Autoregressive Moving Average Model ARMA(,) This section is an introduction to a wide class of models ARMA(p,q) which we will consider in more detail later in this
84 CHAPTER 4. STATIONARY TS MODELS 4.6 Autoregressive Moving Average Model ARMA(,) This section is an introduction to a wide class of models ARMA(p,q) which we will consider in more detail later in this
New bounds for spherical two-distance sets and equiangular lines
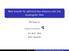 New bounds for spherical two-distance sets and equiangular lines Michigan State University Oct 8-31, 016 Anhui University Definition If X = {x 1, x,, x N } S n 1 (unit sphere in R n ) and x i, x j = a
New bounds for spherical two-distance sets and equiangular lines Michigan State University Oct 8-31, 016 Anhui University Definition If X = {x 1, x,, x N } S n 1 (unit sphere in R n ) and x i, x j = a
The ε-pseudospectrum of a Matrix
 The ε-pseudospectrum of a Matrix Feb 16, 2015 () The ε-pseudospectrum of a Matrix Feb 16, 2015 1 / 18 1 Preliminaries 2 Definitions 3 Basic Properties 4 Computation of Pseudospectrum of 2 2 5 Problems
The ε-pseudospectrum of a Matrix Feb 16, 2015 () The ε-pseudospectrum of a Matrix Feb 16, 2015 1 / 18 1 Preliminaries 2 Definitions 3 Basic Properties 4 Computation of Pseudospectrum of 2 2 5 Problems
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 24/3/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Όλοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα μικρότεροι του 10000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Αν κάπου κάνετε κάποιες υποθέσεις
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Όλοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα μικρότεροι του 10000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Αν κάπου κάνετε κάποιες υποθέσεις
Spherical Coordinates
 Spherical Coordinates MATH 311, Calculus III J. Robert Buchanan Department of Mathematics Fall 2011 Spherical Coordinates Another means of locating points in three-dimensional space is known as the spherical
Spherical Coordinates MATH 311, Calculus III J. Robert Buchanan Department of Mathematics Fall 2011 Spherical Coordinates Another means of locating points in three-dimensional space is known as the spherical
(1) Describe the process by which mercury atoms become excited in a fluorescent tube (3)
 Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Integrals in cylindrical, spherical coordinates (Sect. 15.7)
 Integrals in clindrical, spherical coordinates (Sect. 5.7 Integration in spherical coordinates. Review: Clindrical coordinates. Spherical coordinates in space. Triple integral in spherical coordinates.
Integrals in clindrical, spherical coordinates (Sect. 5.7 Integration in spherical coordinates. Review: Clindrical coordinates. Spherical coordinates in space. Triple integral in spherical coordinates.
CHAPTER 101 FOURIER SERIES FOR PERIODIC FUNCTIONS OF PERIOD
 CHAPTER FOURIER SERIES FOR PERIODIC FUNCTIONS OF PERIOD EXERCISE 36 Page 66. Determine the Fourier series for the periodic function: f(x), when x +, when x which is periodic outside this rge of period.
CHAPTER FOURIER SERIES FOR PERIODIC FUNCTIONS OF PERIOD EXERCISE 36 Page 66. Determine the Fourier series for the periodic function: f(x), when x +, when x which is periodic outside this rge of period.
Bayesian statistics. DS GA 1002 Probability and Statistics for Data Science.
 Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Finite Field Problems: Solutions
 Finite Field Problems: Solutions 1. Let f = x 2 +1 Z 11 [x] and let F = Z 11 [x]/(f), a field. Let Solution: F =11 2 = 121, so F = 121 1 = 120. The possible orders are the divisors of 120. Solution: The
Finite Field Problems: Solutions 1. Let f = x 2 +1 Z 11 [x] and let F = Z 11 [x]/(f), a field. Let Solution: F =11 2 = 121, so F = 121 1 = 120. The possible orders are the divisors of 120. Solution: The
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ
 ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
Congruence Classes of Invertible Matrices of Order 3 over F 2
 International Journal of Algebra, Vol. 8, 24, no. 5, 239-246 HIKARI Ltd, www.m-hikari.com http://dx.doi.org/.2988/ija.24.422 Congruence Classes of Invertible Matrices of Order 3 over F 2 Ligong An and
International Journal of Algebra, Vol. 8, 24, no. 5, 239-246 HIKARI Ltd, www.m-hikari.com http://dx.doi.org/.2988/ija.24.422 Congruence Classes of Invertible Matrices of Order 3 over F 2 Ligong An and
CORDIC Background (2A)
 CORDIC Background 2A Copyright c 20-202 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version.2 or any later
CORDIC Background 2A Copyright c 20-202 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version.2 or any later
Solution to Review Problems for Midterm III
 Solution to Review Problems for Mierm III Mierm III: Friday, November 19 in class Topics:.8-.11, 4.1,4. 1. Find the derivative of the following functions and simplify your answers. (a) x(ln(4x)) +ln(5
Solution to Review Problems for Mierm III Mierm III: Friday, November 19 in class Topics:.8-.11, 4.1,4. 1. Find the derivative of the following functions and simplify your answers. (a) x(ln(4x)) +ln(5
Bounding Nonsplitting Enumeration Degrees
 Bounding Nonsplitting Enumeration Degrees Thomas F. Kent Andrea Sorbi Università degli Studi di Siena Italia July 18, 2007 Goal: Introduce a form of Σ 0 2-permitting for the enumeration degrees. Till now,
Bounding Nonsplitting Enumeration Degrees Thomas F. Kent Andrea Sorbi Università degli Studi di Siena Italia July 18, 2007 Goal: Introduce a form of Σ 0 2-permitting for the enumeration degrees. Till now,
Lifting Entry (continued)
 ifting Entry (continued) Basic planar dynamics of motion, again Yet another equilibrium glide Hypersonic phugoid motion Planar state equations MARYAN 1 01 avid. Akin - All rights reserved http://spacecraft.ssl.umd.edu
ifting Entry (continued) Basic planar dynamics of motion, again Yet another equilibrium glide Hypersonic phugoid motion Planar state equations MARYAN 1 01 avid. Akin - All rights reserved http://spacecraft.ssl.umd.edu
Physical DB Design. B-Trees Index files can become quite large for large main files Indices on index files are possible.
 B-Trees Index files can become quite large for large main files Indices on index files are possible 3 rd -level index 2 nd -level index 1 st -level index Main file 1 The 1 st -level index consists of pairs
B-Trees Index files can become quite large for large main files Indices on index files are possible 3 rd -level index 2 nd -level index 1 st -level index Main file 1 The 1 st -level index consists of pairs
ANSWERSHEET (TOPIC = DIFFERENTIAL CALCULUS) COLLECTION #2. h 0 h h 0 h h 0 ( ) g k = g 0 + g 1 + g g 2009 =?
 Teko Classes IITJEE/AIEEE Maths by SUHAAG SIR, Bhopal, Ph (0755) 3 00 000 www.tekoclasses.com ANSWERSHEET (TOPIC DIFFERENTIAL CALCULUS) COLLECTION # Question Type A.Single Correct Type Q. (A) Sol least
Teko Classes IITJEE/AIEEE Maths by SUHAAG SIR, Bhopal, Ph (0755) 3 00 000 www.tekoclasses.com ANSWERSHEET (TOPIC DIFFERENTIAL CALCULUS) COLLECTION # Question Type A.Single Correct Type Q. (A) Sol least
Every set of first-order formulas is equivalent to an independent set
 Every set of first-order formulas is equivalent to an independent set May 6, 2008 Abstract A set of first-order formulas, whatever the cardinality of the set of symbols, is equivalent to an independent
Every set of first-order formulas is equivalent to an independent set May 6, 2008 Abstract A set of first-order formulas, whatever the cardinality of the set of symbols, is equivalent to an independent
Concrete Mathematics Exercises from 30 September 2016
 Concrete Mathematics Exercises from 30 September 2016 Silvio Capobianco Exercise 1.7 Let H(n) = J(n + 1) J(n). Equation (1.8) tells us that H(2n) = 2, and H(2n+1) = J(2n+2) J(2n+1) = (2J(n+1) 1) (2J(n)+1)
Concrete Mathematics Exercises from 30 September 2016 Silvio Capobianco Exercise 1.7 Let H(n) = J(n + 1) J(n). Equation (1.8) tells us that H(2n) = 2, and H(2n+1) = J(2n+2) J(2n+1) = (2J(n+1) 1) (2J(n)+1)
Solutions to the Schrodinger equation atomic orbitals. Ψ 1 s Ψ 2 s Ψ 2 px Ψ 2 py Ψ 2 pz
 Solutions to the Schrodinger equation atomic orbitals Ψ 1 s Ψ 2 s Ψ 2 px Ψ 2 py Ψ 2 pz ybridization Valence Bond Approach to bonding sp 3 (Ψ 2 s + Ψ 2 px + Ψ 2 py + Ψ 2 pz) sp 2 (Ψ 2 s + Ψ 2 px + Ψ 2 py)
Solutions to the Schrodinger equation atomic orbitals Ψ 1 s Ψ 2 s Ψ 2 px Ψ 2 py Ψ 2 pz ybridization Valence Bond Approach to bonding sp 3 (Ψ 2 s + Ψ 2 px + Ψ 2 py + Ψ 2 pz) sp 2 (Ψ 2 s + Ψ 2 px + Ψ 2 py)
Uniform Convergence of Fourier Series Michael Taylor
 Uniform Convergence of Fourier Series Michael Taylor Given f L 1 T 1 ), we consider the partial sums of the Fourier series of f: N 1) S N fθ) = ˆfk)e ikθ. k= N A calculation gives the Dirichlet formula
Uniform Convergence of Fourier Series Michael Taylor Given f L 1 T 1 ), we consider the partial sums of the Fourier series of f: N 1) S N fθ) = ˆfk)e ikθ. k= N A calculation gives the Dirichlet formula
Variational Wavefunction for the Helium Atom
 Technische Universität Graz Institut für Festkörperphysik Student project Variational Wavefunction for the Helium Atom Molecular and Solid State Physics 53. submitted on: 3. November 9 by: Markus Krammer
Technische Universität Graz Institut für Festkörperphysik Student project Variational Wavefunction for the Helium Atom Molecular and Solid State Physics 53. submitted on: 3. November 9 by: Markus Krammer
Pg The perimeter is P = 3x The area of a triangle is. where b is the base, h is the height. In our case b = x, then the area is
 Pg. 9. The perimeter is P = The area of a triangle is A = bh where b is the base, h is the height 0 h= btan 60 = b = b In our case b =, then the area is A = = 0. By Pythagorean theorem a + a = d a a =
Pg. 9. The perimeter is P = The area of a triangle is A = bh where b is the base, h is the height 0 h= btan 60 = b = b In our case b =, then the area is A = = 0. By Pythagorean theorem a + a = d a a =
Code Breaker. TEACHER s NOTES
 TEACHER s NOTES Time: 50 minutes Learning Outcomes: To relate the genetic code to the assembly of proteins To summarize factors that lead to different types of mutations To distinguish among positive,
TEACHER s NOTES Time: 50 minutes Learning Outcomes: To relate the genetic code to the assembly of proteins To summarize factors that lead to different types of mutations To distinguish among positive,
