Supplementary Appendix
|
|
|
- Αμάλθεια Σπηλιωτόπουλος
- 7 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful Gene-Expression Profiles in Acute Myeloid Leukemia. N Engl J Med 2004;350:
2 Web publication only 1. Supporting Materials and Methods Figure A: Guidelines how to read the Omniviz Correlation View 2. Supporting results Pearson s correlation coefficient analyses using Omniviz with different probe subsets Figure B: 147 probe sets Figure C: 293 probe sets Figure D: 569 probe sets Figure E: 984 probe sets Figure F: 1692 probe sets Figure G: 2856 probe sets Figure H: 5071 probe sets 3. Supporting results assigned clusters (2856 probe sets) Figures Figure I: Southern blot analyses AML patients with cryptic inv(16) Figure J: Expression of MYH11 as determined by Affymetrix GeneChip analyses in 285 cases of AML and controls Figure K: Snapshot of Correlation View showing the AML-M3 t(15;17) patients Figure L: Expression of ETO as determined by Affymetrix GeneChip analyses in 285 cases of AML and controls Tables Cluster Characteristics SAM genes #1 (EVI1) Table A Table A1 #2 (FLT3 ITD) Table B Table B1 #3 Table C Table C1 #4 (cebpα) Table D Table D1 #5 Table E Table E1 #6 (FLT3 ITD) Table F Table F1 #7 Table G Table G1 #8 Table H Table H1 #9 (inv(16)) Table I Table I1 #10 (EVI1) Table J Table J1 #11 Table K Table K1 #12 (t(15;17)) Table L Table L1 #13 (t(8;21)) Table M Table M1 #14 Table N Table N1 #15 (cebpα) Table O Table O1 #16 (11q23) Table P Table P1 Summary of frequencies and percentages of cytogenetic and molecular abnormalities for each of the assigned clusters (Table Q). 4. PAM genes of prognostically important clusters (#13, #12, #9, #16, #10, #4, #15, #4 and #15, and FLT3 ITD) (Table R)
3 FAB Figure A: Correlation View and FAB classification of the cohort of 285 AML patients (2856 probe sets). Correlation (red) between cluster #5 and #16 is indicated by rectangle. Both clusters predominantly consist of AML-M5 and correlate. However, they do form separate clusters. Anticorrelation (blue) for instance between cluster 5 and cluster #13, which merely contains AML-M2, is indicated by the dashed rectangle. Correlation and anti-correlation between every individual (sub)cluster can be extracted from the Correlation View and (sub)clusters can subsequently be assigned, e.g., cluster #6, #7 and #8 (dotted lines). FAB: M0-bright green, M1-green, M2-pink, M3- orange, M4-purple, M5-turquoise, M6-yellow (with number).
4 Figure B: Pearson s correlation coefficient analyses using Omniviz with 147 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
5 Figure C: Pearson s correlation coefficient analyses using Omniviz with 293 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
6 Figure D: Pearson s correlation coefficient analyses using Omniviz with 569 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
7 Figure E: Pearson s correlation coefficient analyses using Omniviz with 984 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
8 Figure F: Pearson s correlation coefficient analyses using Omniviz with 1692 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
9 Figure G: Pearson s correlation coefficient analyses using Omniviz with 2856 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
10 Figure H: Pearson s correlation coefficient analyses using Omniviz with 5071 probe sets. In the Correlation View all 285 patients are plotted versus all 285 AML patients. FAB classification and karyotype based on cytogenetics are depicted in the columns along the original diagonal of the Correlation View (FAB M0-black, M1-green, M2-purple, M3-orange, M4-yellow, M5-turquoise, M6-grey; karyotype: normal-green, inv(16)-yellow, t(8;21)-purple, t(15;17)-orange, 11q23 abnormalities-turquoise, 7(q) abnormalities-red, +8-pink, complex-black, other-grey). FLT3 ITD, FLT3 TKD, N-RAS, K-RAS and cebpα mutations and EVI1 overexpression are depicted in the same set of columns (red bar: positive and green bar: negative).
11 WT WT inv(16) type A inv(16) type A inv(16) type E BamHI - normal allele EcoRI - normal allele Figure I: Southern blot analyses AML patients with cryptic inv(16). Southern blot analyses was carried out with a myosine heavy chain 11 specific probe (NT , nt) on material of AML (WT, no inv(16)), AML with known inv(16) breakpoint (type A and E) and three patients that clustered with all known AML with inv(16) patients in the Correlation View (Figure 1).
12 MYH11 expression 8,0 7,0 6,0 5,0 4,0 3,0 -inv16 +inv16 NBM CD34+ 2,0 1,0 0,0 Figure J: Expression of MYH11 as determined by Affymetrix GeneChip analyses in 285 cases of AML and controls. Expression levels of MYH11 were high in AML patients and inv(16), whereas low levels were detected in the other AML patients, CD34-positive cells and normal bone marrow.
13 FAB karyotype WBC FLT3 ITD FLT3 TKD AML-M3 t(15;17) Figure K: Snapshot of Correlation View showing the AML-M3 t(15;17) patients. FAB M2-purple, M3-orange, M4-yellow. Karyotype: normal-green, t(15;17)-orange, other-grey. The AML-M3 t(15;17) patients are divided into two groups, i.e., low white blood cell count (WBC) and FLT3 ITD 9 negative (green bar) versus high WBC/ FLT3 ITD positive (red bar). Karyotype is based on cytogenetics and WBC is depicted as 10 (cells/l).
14 ETO expression 40,0 35,0 30,0 25,0 20,0 15,0 -t(8;21) +t(8;21) NBM CD34+ 10,0 5,0 0,0 Figure L: Expression of ETO as determined by Affymetrix GeneChip analyses in 285 cases of AML and controls. Expression levels of ETO were high in AML patients and t(8;21), whereas low levels were detected in the other AML patients, CD34-positive cells and normal bone marrow.
15 Table A: Characteristics cluster #1 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 1595 #1 M1 NN #1 M1 NN #1 M1 Complex #1 M1 NN #1 M1 11q23 (t(4;11)) #1 M1 +11/11q23(sMLL) #1 M1 +11/+11/Other #1 M2 NN #1 M5 NN #1 M5 11q23 (t(11;19)) #1 M4 Other/11q23 (t(2;9;11)) #1 M1 11q23 (t(6;11)) #1 M5 11q23 (t(6;11)) #1 M5 NN
16 Table B: Characteristics cluster #2 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 3330 #2 M #2 M1 NN #2 ND NN #2 M4-9q #2 M4 NN #2 M4 t(6;9) #2 M4 NN #2 M5 NN #2 M5 NN #2 M1 NN #2 M5 NN #2 M4 NN #2 M2 NN #2 M4 NN ND 1551 #2 M1 NN #2 M4 NN #2 M
17 Table C: Characteristics cluster #3 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2480 #3 M1 NN #3 M2 NN #3 M1 NN #3 M2 NN #3 M2 NN #3 M1 NN #3 M1 NN #3 M1 NN #3 M1 NN #3 M2 t(9;22) #3 M1 +8/Other #3 M4 NN - ND #3 M4-7/11q #3 M2 t(6;9)/other #3 M5 t(6;9) #3 M1 NN #3 M2 NN #3 M #3 M2 ND
18 Table D: Characteristics cluster #4 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 3327 #4 M1 NN #4 M1-9q #4 M0 Complex #4 M1 NN #4 ND Complex (+8, +11) #4 M1 NN #4 M1 Other #4 M1 NN #4 M1 NN/11q23 (smll) #4 M1 NN #4 M1 NN #4 M1 NN #4 M1 NN #4 M1-9q #4 M1 NN
19 Table E: Characteristics cluster #5 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 3301 #5 M5-5/7(q) #5 M4 NN #5 M5 +8/Other #5 M5 NN - - ND ND #5 M4 ND #5 M5 NN #5 M4 +8/Other #5 M4 NN #5 M4 Complex #5 M #5 M5 Other #5 M5 +8/Other #5 M4 NN #5 M4 +8/Other #5 M5 NN #5 M4 NN #5 M5 ND #5 M5 +8/3q/Other #5 M4 NN #5 M5 NN #5 M5 NN #5 M4 NN #5 ND NN
20 Table E: Characteristics cluster #5 (continued) (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K-RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 3321 #5 M #5 M5 Other #5 M5 NN #5 M4 NN #5 M5 NN/11q23 (smll) #5 M5 NN #5 M5 NN #5 M5 NN #5 M5 NN #5 M5 NN #5 M #5 M5 t(6;9) #5 M5 NN #5 M5 NN #5 M5 NN #5 M5 NN #5 M4 NN #5 M4 NN #5 M5 NN #5 M2 Complex #5 M0 Complex
21 Table F: Characteristics cluster #6 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2683 #6 M2 NN #6 M1 NN #6 M2 NN #6 M1 NN #6 M1 NN #6 M2 NN #6 M1 NN #6 M1 ND
22 Table G: Characteristics cluster #7 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 3310 #7 M2 NN #7 M3 NN #7 M1 NN #7 M1 NN #7 M1 NN #7 M2 NN #7 M2 Other #7 M6 NN #7 M1 NN #7 M6 Other #7 M6 NN #7 M5 NN #7 ND Other #7 M2 +8/Other #7 M2 Complex(3q/+8) #7 M1 NN #7 M2 NN #7 M3 ND
23 Table H: Characteristics cluster #8 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2223 #8 M #8 M5 Complex (-7(q)/+8) #8 M2 Complex (11q23 (t(8;11)), -5, 3q) #8 ND +11/Other #8 M2 NN #8 ND +8,-7(q) #8 M2 NN #8 M2 inv7(q)/other #8 M #8 M2 NN #8 M2 Other #8 M0 Other #8 M2 NN
24 Table I: Characteristics cluster #9 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, BP:inv(16) breakpoint, RT: real-time PCR for CBFβ-MYH11 (Primer CBFβ 5 -AAGACTGGATGGTATGGGCTGT-3 (sense), Primer 126REV 5 -CAGGGCCCGCTTGGA-3 (antisense), Probe CBFβ 6- FAM 5 -TGGAGTTTGATGAGGAGCGAGCCC-3 TAMRA); FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K-RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype BP RT FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 3277 #9 M1 idt(16) A #9 M4 idt(16) A #9 M4 idt(16)/-7(q) A #9 M5 idt(16) A #9 M4 idt(16) A #9 M4 idt(16) A #9 M4 NN A #9 M5 idt(16) A #9 M4 idt(16) A #9 M4 NN A #9 M4 idt(16) A #9 M4 idt(16) D #9 M4 idt(16) A #9 M4 idt(16) A #9 M5-7(q) A #9 M4 idt(16)/+8 A #9 M4 idt(16) A ND #9 M4 idt(16) A #9 ND idt(16) A #9 M4 idt(16) A #9 M4 NN A #9 M2 idt(16)/+8 A #9 M4 idt(16) A
25 Table J: Characteristics cluster #10 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2661 #10 M4 ND #10 M #10 M5-7/3q #10 M2-7(q) #10 M5 Other #10 M0 Other #10 M #10 M1 t(9;22) #10 M #10 M #10 M0 Other #10 M0-7/3q #10 M5 ND #10 M1-7(q) #10 M1 Other #10 ND ND #10 M #10 M2 NN #10 M #10 M1 NN #10 M1 Other #10 M1 Other
26 Table K: Characteristics cluster #11 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2209 #11 M4 Other #11 M4 NN #11 M5 Other #11 M5 NN #11 M2 NN #11 M1 NN #11 M5 NN #11 ND NN #11 M5 NN
27 Table L: Characteristics cluster #12 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR for PML-RARα (Primer PML3-for 5 -CCCCAGGAGCCCCGT-3 (sense), Primer PML-kbr 5 -CCTGCAGGACCTCAGCTCTT-3 (sense), Primer RAR4-rev 5 - AAAGCAAGGCTTGTAGATGCG-3 (antisense), Probe RARA 6-FAM 5 -AGTGCCCAGCCCTCCCTCGC-3 TAMRA); FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K-RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype RT FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2466 #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17)/other #12 M3 t(15;17)/other #12 M3 t(15;17) #12 M3 t(15;17)/other #12 M2 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 t(15;17) #12 M3 Other* #12 M4 t(15;17)/other #12 M3 t(15;17)/ *Full karyotype of patient 322: 46,XX, add(12)(p1?3).
28 Table M: Characteristics cluster #13 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR for AML1-ETO (Primer 821 For 5 -TCACTCTGACCATCACTGTCTTCA-3 (sense), Primer 821 Rev 5 -ATTGTGGAGTGCTTCTCAGTACGAT -3 (antisense), Probe ETO 6- FAM 5 -ACCCACCGCAAGTCGCCACCT -3 TAMRA); FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K-RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype RT FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2243 #13 M2 t(+8;21)/other #13 M4 t(+8;21) #13 M2 t(+8;21) #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other #13 M2 t(+8;21) #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other #13 M2 t(+8;21)/+8/other #13 M2 t(+8;21)/other #13 M2 t(+8;21) #13 M2 t(+8;21) #13 M2 t(+8;21)/other #13 M2 t(+8;21) #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other #13 M1 t(+8;21) #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other #13 M2 t(+8;21)/other
29 Table N: Characteristics cluster #14 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2536 #14 ND ND #14 M2 ND #14 M2 +8/Other #14 M2 11q23 (ND) #14 M2-5(q) #14 M1 Complex(-5/-7/+8) #14 M4 Complex #14 M #14 M2 NN #14 M5 NN
30 Table O: Characteristics cluster #15 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2767 #15 M1 ND #15 M4 NN #15 M1 NN #15 M2 NN #15 M2 Other #15 M2 NN #15 M2 NN #15 M1-7/Other
31 Table P: Characteristics cluster #16 (Patient: patient number, Cluster: cluster number (2856 probe sets); FAB: FAB subtype of AML; Karyotype: t(15;17), t(8;21), inv(16)/t(16;16),+8,+11,+21,-5(q),-7(q),t(9;22),3q abnormalities, 11q23 abnormalities (translocation/self fusion (smll)), complex(abnormalities involved) (>3abnormalities) and normal karyotype (NN) are indicated, RT: real-time PCR; FLT3 ITD: internal tandem duplication in FLT3; FLT3 TKD: tyrosine kinase domain mutation in FLT3; N- or K-RAS: mutation in codon 12,13 or 61 of N- or K- RAS; EVI1: EVI1 overexpression; CEPBA: mutation in CEBPA, ND: not determined). Patient Cluster FAB Karyotype FLT3 ITD FLT3 TKD N-RAS K-RAS EVI1 CEBPA 2225 #16 M4 NN #16 M5 Other #16 M5 Other #16 M5 +8/11q23 (t(11;19) #16 M5 11q23 (t(9;11)) #16 M5 Other/11q23 (t(9;11)) #16 M5 11q23 (t(9;11)) #16 M5 Other #16 M5 NN #16 M1 NN #16 M5 11q23 (t(9;11))/
32 Table Q: Frequency and percentage of cytogenetic and molecular abnormalities of all AML patients within each of the assigned clusters. All patients with a specific abnormality were considered, irrespective of the presence of additional abnormalities (NC: patients not assigned to any of the 16 clusters). Cluster #1 #2 #3 #4 #5 #6 #7 #8 #9 #10 #11 #12 #13 #14 #15 #16 NC total Patients in cluster Cytogenetics t(15;17) 18 (95) 18 (6) t(8;21) 22 (100) 22 (8) inv(16)/t(16;16) 19 (83) 19 (7) +8 2 (12) 1 (5) 1 (7) 7 (16) 2 (11) 2 (15) 2 (9) 1 (5) 1 (5) 1 (5) 3 (30) 1 (9) 2 (15) 26 (9) (14) 1 (7) 1 (2) 1 (8) 1 (5) 1 (8) 7 (2) (5) 1 (8) 2 (1) -5 1 (8) 1 (5) 1 (10) 3 (1) -5(q) 1 (10) 1 (<1) -7 1 (5) 1 (2) 1 (8) 5 (23) 1 (10) 1 (13) 1 (9) 2 (15) 13 (5) -7(q) 3 (23) 2 (9) 2 (9) 7 (2) 3q 1 (2) 1 (6) 1 (8) 2 (9) 1 (8) 6 (2) t(6;9) 1 (6) 2 (11) 1 (2) 4 (1) t(9;22) 1 (5) 1 (5) 2 (1) t(11q23) 6 (43) 1 (5) 2 (13) 1 (2) 1 (8) 1 (10) 5 (45) 2 (15) 19 (7) complex (>3 abn.) 1 (7) 2 (13) 3 (7) 1 (6) 2 (15) 2 (20) 11 (4) other 2 (14) 1 (6) 2 (11) 3 (20) 7 (16) 4 (22) 4 (31) 6 (27) 2 (22) 4 (21) 15 (68) 1 (10) 2 (25) 4 (36) 3 (23) 60 (21) normal 6 (43) 13 (76) 13 (68) 10 (67) 27 (61) 7 (88) 12 (67) 4 (31) 3 (13) 2 (9) 7 (78) 2 (20) 5 (63) 3 (27) 5 (38) 119 (42) ND 2 (5) 1 (13) 1 (6) 3 (14) 2 (20) 1 (13) 10 (4) Cluster #1 #2 #3 #4 #5 #6 #7 #8 #9 #10 #11 #12 #13 #14 #15 #16 NC total Patients in cluster Molecular markers FLT3-ITD 2 (14) 14 (82) 10 (53) 1 (7) 14 (32) 8 (100) 4 (22) 4 (18) 1 (11) 6 (32) 1 (5) 3 (30) 2 (25) 8 (62) 78 (27) FLT3-TKD 3 (18) 3 (16) 6 (14) 1 (13) 6 (26) 1 (5) 3 (33) 5 (26) 1 (5) 2 (20) 1 (13) 1 (9) 33 (12) N-RAS 1 (7) 4 (9) 1 (8) 8 (35) 3 (14) 2 (22) 3 (14) 1 (10) 1 (13) 2 (15) 26 (9) K-RAS 1 (7) 4 (9) 2 (25) 1 (4) 1 (5) 9 (3) EVI1 5 (36) 2 (11) 2 (5) 2 (11) 10 (45) 2 (15) 23 (8) CEBPA 1 (6) 8 (53) 1 (2) 1 (5) 1 (5) 5 (63) 17 (6)
33 Table A1: Top40 genes cluster #1 Probe Set ID Gene Symbol Locus Link Accession Score SAM _at LOC NM_ ,09 1, _at KCNA NM_ ,68 1, _at GPR AL ,18 1, _s_at TRPS NM_ ,95 1, _s_at BHLHB AB ,63 1, _at EVI BE ,40 1, _s_at KIAA NM_ ,96 1, _at IGHM 3507 X ,85 1, _at MMRN NM_ ,72 1, _at MEF2C 4208 N ,59 1, _at ATP10A AB ,41 1, _s_at FHL U ,37 1, _x_at PCDHGC NM_ ,29 1, _at KIF AA ,25 1, _s_at LTBP NM_ ,21 1, _s_at PCDHGC AK ,20 1, _at SPIB 6689 NM_ ,15 1, _s_at SOCS AB ,12 1, _x_at PCDHGC AF ,11 1, _at AF ,09 1, _s_at MEF2C 4208 N ,08 1, _s_at BLNK NM_ ,05 1, _s_at DPP M ,03 1, _s_at AI ,01 1, _s_at PROM NM_ ,97 1, _at SOCS NM_ ,95 1, _s_at SLC38A NM_ ,87 1, _at BMI1 648 NM_ ,86 1, _x_at FHL AF ,83 1, _s_at IRF NM_ ,77 1, _s_at SPAG AI ,77 1, _s_at SLC2A AA ,76 1, _s_at NR4A AI ,80 1, _s_at NR4A S ,84 1, _s_at SLC2A AL ,85 1, _x_at NR4A NM_ ,85 1, _x_at SLC2A NM_ ,91 1, _x_at JUN 3725 BG ,92 1, _s_at ATF3 467 NM_ ,11 1,96 q-value SAM (%)
34 Table B1: Top 40 genes cluster #2 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _s_at GLI NM_ ,30 1, _at IL2RA 3559 NM_ ,15 1, _s_at IL2RA 3559 K ,24 1, _at TRPC AI ,44 1, _at PLS NM_ ,34 1, _at PLA2G4A 5321 M ,31 1, _at TRIM NM_ ,23 1, _s_at PTP4A NM_ ,01 1, _at DOCK NM_ ,48 1, _s_at KCNK NM_ ,29 1, _at LPIN D ,05 1, _at RhoGAP NM_ ,82 1, _at CCL NM_ ,69 1, _s_at GPR NM_ ,41 1, _s_at GOLGIN AI ,37 1, _at HOXB NM_ ,12 1, _at GPR AL ,01 1, _at GRB D ,99 1, _x_at GOLGIN AF ,97 1, _s_at LAPTM4B AW ,95 1, _s_at GUCY1A AI ,95 1, _at PIM M ,94 1, _s_at SCHIP NM_ ,89 1, _at HOXA NM_ ,74 1, _x_at GOLGIN AF ,70 1, _s_at JAG1 182 U ,68 1, _s_at CLU 1191 M ,60 1, _x_at V ,62 1, _x_at HBA AF ,67 1, _x_at AF ,71 1, _x_at HBB 3043 M ,71 1, _x_at HBA T ,72 1, _x_at HBB 3043 AF ,72 1, _x_at HBA BC ,75 1, _x_at HBA NM_ ,83 1, _s_at VIL J ,91 1, _x_at HBA AF ,96 1, _at PDE3B 5140 NM_ ,29 1, _s_at AI ,39 1, _s_at MAP4K NM_ ,87 1,04
35 Table C1: Top40 genes cluster #3 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _at SCN9A 6335 NM_ ,09 0, _at GAS NM_ ,63 0, _at CCL NM_ ,56 0, _at PLS NM_ ,94 0, _s_at COL4A AW ,51 0, _s_at LAPTM4B AW ,09 0, _at RhoGAP NM_ ,07 0, _s_at LAPTM4B NM_ ,05 0, _at QPRT NM_ ,04 0, _x_at ANPEP 9053 T ,84 0, _at LOC NM_ ,81 0, _at SMC4L AL ,81 0, _at SH2D1A 4068 AF ,74 0, _at ADCY2 108 AU ,53 0, _s_at ENPP AW ,48 0, _at TRIM NM_ ,42 0, _s_at LAPTM4B T ,41 0, _s_at GPR NM_ ,28 0, _at MAP T ,28 0, _s_at MAP AJ ,23 0, _s_at FLJ NM_ ,20 0, _s_at SCDGF-B NM_ ,05 0, _at DOCK NM_ ,03 0, _s_at HFL X ,00 0, _s_at SMC4L NM_ ,00 0, _x_at TNFRSF AJ ,96 0, _s_at SEPP NM_ ,94 0, _x_at PBXIP NM_ ,92 0, _at NET AW ,85 0, _at C14orf NM_ ,85 0, _s_at KIAA NM_ ,88 0, _at IL17R NM_ ,95 0, _at IL6ST 3572 AL ,03 0, _at SERPINB NM_ ,11 0, _x_at BLVRA 644 NM_ ,71 0, _at RAP2A 5911 AI ,94 0, _s_at FCGRT 2217 NM_ ,10 0, _x_at BLVRA 644 BC ,18 0, _s_at AGTPBP NM_ ,15 0, _at AIM1 202 U ,19 0,21
36 Table D1: Top40 genes cluster #4 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _at AV ,34 0, _s_at 6964 X ,01 0, _s_at B4GALT NM_ ,59 0, _at 6964 AW ,85 0, _x_at UGT2B AF ,60 0, _at FLJ NM_ ,57 0, _s_at 6964 X ,55 0, _s_at CD7 924 NM_ ,22 0, _x_at CD7 924 AI ,04 0, _at IGFBP AW ,85 0, _s_at C18orf1 753 NM_ ,65 0, _at ZNFN1A NM_ ,27 0, _s_at ABCB AF ,90 0, _at B4GALT AF ,66 0, _s_at TRIM AJ ,44 0, _at ABCB AF ,40 0, _s_at P2RX U ,36 0, _x_at PMS2L D ,20 0, _at HPS AL ,16 0, _at PGDS NM_ ,79 0, _at TM4SF NM_ ,79 0, _at FZD NM_ ,63 0, _at BG ,50 0, _at ITGA NM_ ,49 0, _at ITGA BG ,37 0, _at FLJ NM_ ,51 0, _at CYFIP BC ,75 0, _s_at KLF NM_ ,95 0, _at IL4R 3566 NM_ ,96 0, _s_at DF 1675 NM_ ,98 0, _at CAPN2 824 M ,08 0, _s_at CSDA 8531 AL ,13 0, _at LRP NM_ ,19 0, _at RAB NM_ ,25 0, _s_at NDFIP NM_ ,98 0, _at C8FW NM_ ,41 0, _at TUBB BC ,60 0, _x_at CTNNA NM_ ,35 0, _s_at CTNNA AI ,70 0, _x_at CTNNA D ,91 0,11
37 Table E1: Top40 genes cluster #5 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _s_at EPB41L NM_ ,03 0, _s_at LILRB NM_ ,91 0, _s_at EPB41L BC ,65 0, _at SIRPB NM_ ,55 0, _at PILRA NM_ ,93 0, _at CAMK NM_ ,41 0, _at DKFZp434L NM_ ,11 0, _at EPB41L AI ,04 0, _s_at UBE2D AL ,87 0, _s_at VDR 7421 NM_ ,69 0, _s_at STS 412 AU ,64 0, _s_at SIGLEC NM_ ,61 0, _at PTAFR 5724 D ,55 0, _s_at CSPG BF ,07 0, _at PHT NM_ ,04 0, _at TLR NM_ ,94 0, _x_at LILRB AF ,91 0, _s_at PILRA AJ ,71 0, _s_at STS 412 AU ,70 0, _s_at ECGF NM_ ,70 0, _at LILRB AF ,70 0, _x_at HNMT 3176 BC ,69 0, _s_at MGC NM_ ,67 0, _s_at KCNQ NM_ ,66 0, _at IFNGR NM_ ,58 0, _at CARD NM_ ,53 0, _x_at LILRB NM_ ,46 0, _at CD BG ,21 0, _s_at CD NM_ ,15 0, _s_at STS 412 NM_ ,05 0, _at GNS 2799 AW ,03 0, _at RASSF AF ,00 0, _s_at MAFB 9935 NM_ ,99 0, _at ASM3A AA ,96 0, _x_at LILRB AF ,91 0, _x_at LILRB NM_ ,90 0, _s_at PSAP 5660 M ,89 0, _s_at CCR NM_ ,87 0, _s_at EPHB D ,85 0, _at TFEB 7942 AI ,81 0,05
38 Table F1: Top40 genes cluster #6 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _s_at AI ,39 0, _at AI ,99 0, _s_at SNCAIP 9627 NM_ ,81 0, _at FTO U ,51 0, _at MGC AK ,40 0, _at GPC NM_ ,34 0, _s_at GPC AF ,25 0, _at DKFZP564M AK ,56 0, _s_at PLXNB AV ,42 0, _s_at FLJ NM_ ,93 0, _at FLJ NM_ ,80 0, _at ADCY2 108 AU ,78 0, _at FLJ NM_ ,78 0, _s_at LTBP AI ,64 0, _at FOXF NM_ ,60 0, _at VLCS-H NM_ ,31 0, _at FOXC AU ,23 0, _x_at SMC4L AK ,19 0, _s_at DPYSL NM_ ,18 0, _s_at HOXB NM_ ,17 0, _at NM_ ,16 0, _s_at DSC BF ,16 0, _at BITE NM_ ,13 0, _s_at HFL X ,12 0, _at NM_ ,08 0, _at CAT 847 AU ,04 0, _x_at DSC NM_ ,01 0, _s_at LTBP NM_ ,97 0, _at BF ,61 0, _at BST2 684 NM_ ,55 0, _x_at HLA-DRB AJ ,56 0, _at CD K ,58 0, _at PECAM AW ,62 0, _s_at HLA-DRA 3122 M ,68 0, _at HLA-DPA M ,84 0, _s_at KIAA AK ,87 0, _at XPA 7507 NM_ ,10 0, _x_at HIG NM_ ,41 0, _s_at RGS NM_ ,69 0, _at CORO1A U ,97 0,85
39 Table G1: Top40 genes cluster #7 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _s_at TPM NM_ ,29 0, _at GYPE 2996 NM_ ,28 0, _x_at PLAB 9518 AF ,76 0, _at TRIM X ,56 0, _s_at ANK1 286 NM_ ,78 0, _s_at DNAJC AV ,68 0, _s_at RHAG 6005 AF ,40 0, _x_at MYL X ,18 0, _x_at MYL M ,16 0, _x_at ANK1 286 M ,09 0, _s_at SLC6A NM_ ,08 0, _at TNS 7145 AF ,98 0, _at RAP1GA NM_ ,94 0, _x_at EPB AF ,93 0, _at HBZ 3050 NM_ ,90 0, _x_at RHAG 6005 AF ,88 0, _x_at ANK1 286 NM_ ,84 0, _x_at ANK1 286 NM_ ,83 0, _at MAP NM_ ,71 0, _s_at SPTB 6710 NM_ ,70 0, _x_at ANK1 286 NM_ ,70 0, _at KCNH NM_ ,67 0, _x_at MYL AF ,65 0, _x_at RHD 6007 AF ,64 0, _x_at SLC6A U ,61 0, _s_at C5orf NM_ ,60 0, _at N ,60 0, _x_at X ,53 0, _at RHAG 6005 NM_ ,51 0, _x_at SLC6A AW ,48 0, _at KEL 3792 NM_ ,47 0, _s_at TAL X ,42 0, _s_at OSBP NM_ ,37 0, _s_at DKFZP434C AK ,27 0, _s_at EPB NM_ ,24 0, _s_at ANK1 286 AI ,21 0, _at SLC2A NM_ ,20 0, _s_at SELENBP NM_ ,18 0, _s_at MSCP NM_ ,13 0, _at CLCN NM_ ,12 0,11
40 Table H1: Top40 genes cluster #8 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _at RIS BF ,86 0, _s_at CDH1 999 NM_ ,12 0, _at ABCG AF ,01 0, _at OPTN AV ,88 0, _at LU 4059 X ,45 0, _at PBX BF ,14 0, _s_at ARHGEF NM_ ,95 0, _x_at RHD 6007 L ,72 0, _x_at ANK1 286 M ,53 0, _s_at OSBP NM_ ,53 0, _at CDC42BPA 8476 NM_ ,44 0, _s_at C5orf NM_ ,42 0, _at MXI NM_ ,29 0, _s_at GYPA 2993 BC ,22 0, _x_at ANK1 286 NM_ ,20 0, _s_at ALS2CR NM_ ,10 0, _at AA ,06 0, _x_at L ,04 0, _at GYPA 2993 NM_ ,04 0, _at SLC6A NM_ ,03 0, _s_at TAL X ,98 0, _at CDC42BPA 8476 NM_ ,96 0, _x_at GYPA 2993 U ,95 0, _at TNS 7145 AF ,94 0, _s_at U ,90 0, _s_at OPTN NM_ ,89 0, _at WDR NM_ ,86 0, _x_at GYPE 2996 U ,84 0, _s_at ALS2CR AV ,84 0, _x_at RHCE 6006 X ,81 0, _s_at SNCA 6622 NM_ ,80 0, _x_at ANK1 286 NM_ ,78 0, _x_at SLC6A AW ,78 0, _x_at RHD 6007 AF ,77 0, _at TM4SF AF ,75 0, _s_at DJ473B NM_ ,74 0, _s_at SELENBP NM_ ,70 0, _r_at C5orf H ,70 0, _x_at ANK1 286 NM_ ,69 0, _at FECH 2235 AU ,66 0,17
41 Table I1: Top40 genes cluster #9 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _x_at MYH NM_ ,02 0, _x_at MYH NM_ ,72 0, _at CLIPR AL ,92 0, _at ST NM_ ,69 0, _at NRP BE ,71 0, _at CLECSF NM_ ,32 0, _s_at PAPSS AF ,04 0, _s_at PAPSS AW ,73 0, _at CD1C 911 NM_ ,82 0, _s_at COLEC NM_ ,69 0, _s_at MSLN NM_ ,36 0, _s_at CHI3L M ,06 0, _at FLJ NM_ ,59 0, _s_at CRA NM_ ,49 0, _at CHI3L M ,07 0, _s_at AK NM_ ,88 0, _s_at ICAM NM_ ,76 0, _at Z39IG NM_ ,23 0, _s_at SPARC 6678 NM_ ,18 0, _at TGFBI 7045 NM_ ,99 0, _at RPS6KA AI ,82 0, _at NT5E 4907 NM_ ,67 0, _at MN NM_ ,24 0, _at SDR NM_ ,92 0, _at LOC AU ,85 0, _s_at FCGR2B 2213 M ,82 0, _at CGI NM_ ,45 0, _at PTPRM 5797 NM_ ,25 0, _s_at RUNX3 864 NM_ ,25 0, _s_at CD NM_ ,33 0, _s_at NDE NM_ ,42 0, _at EMU AL ,56 0, _s_at RUNX3 864 AA ,85 0, _s_at MGLL BC ,01 0, _at FLJ NM_ ,61 0, _s_at CBFB 865 AF ,73 0, _s_at CHST NM_ ,82 0, _s_at CYLN BC ,24 0, _s_at CBFB 865 NM_ ,01 0, _at CD NM_ ,28 0,18
42 Table J1: Top40 genes cluster #10 Probe Set ID Gene Symbol Locus Link Accession Score SAM q-value SAM (%) _at FLJ NM_ ,59 0, _s_at CRIM BG ,82 0, _at FLJ AI ,75 0, _at NRLN AL ,99 0, _s_at SPTBN NM_ ,75 0, _at FLJ AL ,75 0, _at AI ,74 0, _at SETBP NM_ ,63 0, _at F2RL BE ,53 0, _at PPP1R16B AB ,52 0, _s_at LOC BC ,51 0, _at GDAP1L BF ,43 0, _s_at SCAM NM_ ,42 0, _s_at TPM NM_ ,21 0, _at RBPMS D ,19 0, _s_at RBPMS NM_ ,14 0, _at PRKD AF ,14 0, _at SPON AB ,12 0, _at PIK3C2B 5287 NM_ ,11 0, _at PPP1R16B AB ,09 0, _at MN NM_ ,03 0, _at GNAI AL ,02 0, _at C14orf NM_ ,91 0, _at P164RHOGEF 9828 NM_ ,89 0, _x_at SPTBN NM_ ,88 0, _at IGHM 3507 X ,86 0, _s_at SPON AB ,74 0, _s_at MLLT AV ,59 0, _at EEF1A NM_ ,57 0, _s_at FLNB 2317 M ,40 0, _at CD NM_ ,29 0, _at NPDC NM_ ,25 0, _s_at RBPMS D ,21 0, _s_at BAALC NM_ ,11 0, _s_at BCL7A 605 AI ,05 0, _s_at SPTBN BE ,93 0, _at RNASE NM_ ,00 0, _at C3AR1 719 U ,34 0, _s_at DF 1675 NM_ ,63 0, _s_at AZU1 566 NM_ ,95 0,21
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
department listing department name αχχουντσ ϕανε βαλικτ δδσϕηασδδη σδηφγ ασκϕηλκ τεχηνιχαλ αλαν ϕουν διξ τεχηνιχαλ ϕοην µαριανι
 She selects the option. Jenny starts with the al listing. This has employees listed within She drills down through the employee. The inferred ER sttricture relates this to the redcords in the databasee
She selects the option. Jenny starts with the al listing. This has employees listed within She drills down through the employee. The inferred ER sttricture relates this to the redcords in the databasee
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Example Sheet 3 Solutions
 Example Sheet 3 Solutions. i Regular Sturm-Liouville. ii Singular Sturm-Liouville mixed boundary conditions. iii Not Sturm-Liouville ODE is not in Sturm-Liouville form. iv Regular Sturm-Liouville note
Example Sheet 3 Solutions. i Regular Sturm-Liouville. ii Singular Sturm-Liouville mixed boundary conditions. iii Not Sturm-Liouville ODE is not in Sturm-Liouville form. iv Regular Sturm-Liouville note
derivation of the Laplacian from rectangular to spherical coordinates
 derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
the total number of electrons passing through the lamp.
 1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Code Breaker. TEACHER s NOTES
 TEACHER s NOTES Time: 50 minutes Learning Outcomes: To relate the genetic code to the assembly of proteins To summarize factors that lead to different types of mutations To distinguish among positive,
TEACHER s NOTES Time: 50 minutes Learning Outcomes: To relate the genetic code to the assembly of proteins To summarize factors that lead to different types of mutations To distinguish among positive,
3.4 SUM AND DIFFERENCE FORMULAS. NOTE: cos(α+β) cos α + cos β cos(α-β) cos α -cos β
 3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
EPL 603 TOPICS IN SOFTWARE ENGINEERING. Lab 5: Component Adaptation Environment (COPE)
 EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
SMD Wire Wound Ferrite Chip Inductors - LS Series. LS Series. Product Identification. Shape and Dimensions / Recommended Pattern LS0402/0603/0805/1008
 SMD Wire Wound Ferrite Chip Inductors - LS Series LS Series LS Series is the newest in open type ferrite wire wound chip inductors. The wire wound ferrite construction supports higher SRF, lower DCR and
SMD Wire Wound Ferrite Chip Inductors - LS Series LS Series LS Series is the newest in open type ferrite wire wound chip inductors. The wire wound ferrite construction supports higher SRF, lower DCR and
ΚΥΠΡΙΑΚΟΣ ΣΥΝΔΕΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY 21 ος ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ Δεύτερος Γύρος - 30 Μαρτίου 2011
 Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ Πτυχιακή εργασία ΓΝΩΣΕΙΣ ΚΑΙ ΣΤΑΣΕΙΣ ΝΟΣΗΛΕΥΤΩΝ ΠΡΟΣ ΤΟΥΣ ΦΟΡΕΙΣ ΜΕ ΣΥΝΔΡΟΜΟ ΕΠΙΚΤΗΤΗΣ ΑΝΟΣΟΑΝΕΠΑΡΚΕΙΑΣ (AIDS) Αλέξης Δημήτρη Α.Φ.Τ: 20085675385 Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ Πτυχιακή εργασία ΓΝΩΣΕΙΣ ΚΑΙ ΣΤΑΣΕΙΣ ΝΟΣΗΛΕΥΤΩΝ ΠΡΟΣ ΤΟΥΣ ΦΟΡΕΙΣ ΜΕ ΣΥΝΔΡΟΜΟ ΕΠΙΚΤΗΤΗΣ ΑΝΟΣΟΑΝΕΠΑΡΚΕΙΑΣ (AIDS) Αλέξης Δημήτρη Α.Φ.Τ: 20085675385 Λεμεσός
[1] P Q. Fig. 3.1
![[1] P Q. Fig. 3.1 [1] P Q. Fig. 3.1](/thumbs/79/80362156.jpg) 1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
EE512: Error Control Coding
 EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
Supporting Information
 Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
Τ.Ε.Ι. ΔΥΤΙΚΗΣ ΜΑΚΕΔΟΝΙΑΣ ΠΑΡΑΡΤΗΜΑ ΚΑΣΤΟΡΙΑΣ ΤΜΗΜΑ ΔΗΜΟΣΙΩΝ ΣΧΕΣΕΩΝ & ΕΠΙΚΟΙΝΩΝΙΑΣ
 Τ.Ε.Ι. ΔΥΤΙΚΗΣ ΜΑΚΕΔΟΝΙΑΣ ΠΑΡΑΡΤΗΜΑ ΚΑΣΤΟΡΙΑΣ ΤΜΗΜΑ ΔΗΜΟΣΙΩΝ ΣΧΕΣΕΩΝ & ΕΠΙΚΟΙΝΩΝΙΑΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η προβολή επιστημονικών θεμάτων από τα ελληνικά ΜΜΕ : Η κάλυψή τους στον ελληνικό ημερήσιο τύπο Σαραλιώτου
Τ.Ε.Ι. ΔΥΤΙΚΗΣ ΜΑΚΕΔΟΝΙΑΣ ΠΑΡΑΡΤΗΜΑ ΚΑΣΤΟΡΙΑΣ ΤΜΗΜΑ ΔΗΜΟΣΙΩΝ ΣΧΕΣΕΩΝ & ΕΠΙΚΟΙΝΩΝΙΑΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η προβολή επιστημονικών θεμάτων από τα ελληνικά ΜΜΕ : Η κάλυψή τους στον ελληνικό ημερήσιο τύπο Σαραλιώτου
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
Reminders: linear functions
 Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Potential Dividers. 46 minutes. 46 marks. Page 1 of 11
 Potential Dividers 46 minutes 46 marks Page 1 of 11 Q1. In the circuit shown in the figure below, the battery, of negligible internal resistance, has an emf of 30 V. The pd across the lamp is 6.0 V and
Potential Dividers 46 minutes 46 marks Page 1 of 11 Q1. In the circuit shown in the figure below, the battery, of negligible internal resistance, has an emf of 30 V. The pd across the lamp is 6.0 V and
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
Homework 3 Solutions
 Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
ΑΚΑ ΗΜΙΑ ΕΜΠΟΡΙΚΟΥ ΝΑΥΤΙΚΟΥ ΜΑΚΕ ΟΝΙΑΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ
 ΑΚΑ ΗΜΙΑ ΕΜΠΟΡΙΚΟΥ ΝΑΥΤΙΚΟΥ ΜΑΚΕ ΟΝΙΑΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΘΕΜΑ :ΤΥΠΟΙ ΑΕΡΟΣΥΜΠΙΕΣΤΩΝ ΚΑΙ ΤΡΟΠΟΙ ΛΕΙΤΟΥΡΓΙΑΣ ΣΠΟΥ ΑΣΤΡΙΑ: ΕΥΘΥΜΙΑ ΟΥ ΣΩΣΑΝΝΑ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ : ΓΟΥΛΟΠΟΥΛΟΣ ΑΘΑΝΑΣΙΟΣ 1 ΑΚΑ
ΑΚΑ ΗΜΙΑ ΕΜΠΟΡΙΚΟΥ ΝΑΥΤΙΚΟΥ ΜΑΚΕ ΟΝΙΑΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΘΕΜΑ :ΤΥΠΟΙ ΑΕΡΟΣΥΜΠΙΕΣΤΩΝ ΚΑΙ ΤΡΟΠΟΙ ΛΕΙΤΟΥΡΓΙΑΣ ΣΠΟΥ ΑΣΤΡΙΑ: ΕΥΘΥΜΙΑ ΟΥ ΣΩΣΑΝΝΑ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ : ΓΟΥΛΟΠΟΥΛΟΣ ΑΘΑΝΑΣΙΟΣ 1 ΑΚΑ
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 6/5/2006
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα είναι μικρότεροι το 1000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Διάρκεια: 3,5 ώρες Καλή
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα είναι μικρότεροι το 1000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Διάρκεια: 3,5 ώρες Καλή
SCHOOL OF MATHEMATICAL SCIENCES G11LMA Linear Mathematics Examination Solutions
 SCHOOL OF MATHEMATICAL SCIENCES GLMA Linear Mathematics 00- Examination Solutions. (a) i. ( + 5i)( i) = (6 + 5) + (5 )i = + i. Real part is, imaginary part is. (b) ii. + 5i i ( + 5i)( + i) = ( i)( + i)
SCHOOL OF MATHEMATICAL SCIENCES GLMA Linear Mathematics 00- Examination Solutions. (a) i. ( + 5i)( i) = (6 + 5) + (5 )i = + i. Real part is, imaginary part is. (b) ii. + 5i i ( + 5i)( + i) = ( i)( + i)
Second Order Partial Differential Equations
 Chapter 7 Second Order Partial Differential Equations 7.1 Introduction A second order linear PDE in two independent variables (x, y Ω can be written as A(x, y u x + B(x, y u xy + C(x, y u u u + D(x, y
Chapter 7 Second Order Partial Differential Equations 7.1 Introduction A second order linear PDE in two independent variables (x, y Ω can be written as A(x, y u x + B(x, y u xy + C(x, y u u u + D(x, y
 Problem 7.19 Ignoring reflection at the air soil boundary, if the amplitude of a 3-GHz incident wave is 10 V/m at the surface of a wet soil medium, at what depth will it be down to 1 mv/m? Wet soil is
Problem 7.19 Ignoring reflection at the air soil boundary, if the amplitude of a 3-GHz incident wave is 10 V/m at the surface of a wet soil medium, at what depth will it be down to 1 mv/m? Wet soil is
svari Real-time RT-PCR RSV
 19 39-43 12 Real-time RS svarireal-time RSV subgroup HMPV RSV N Real-time 100 100 Real-time RSV subgroup RSV HMPV F Real-time 55.8 95.5 genotype A2 B1 TaqMan svari InfV RS RSV HMPV HRV PIV RSV HMPV RSV
19 39-43 12 Real-time RS svarireal-time RSV subgroup HMPV RSV N Real-time 100 100 Real-time RSV subgroup RSV HMPV F Real-time 55.8 95.5 genotype A2 B1 TaqMan svari InfV RS RSV HMPV HRV PIV RSV HMPV RSV
Section 1: Listening and responding. Presenter: Niki Farfara MGTAV VCE Seminar 7 August 2016
 Section 1: Listening and responding Presenter: Niki Farfara MGTAV VCE Seminar 7 August 2016 Section 1: Listening and responding Section 1: Listening and Responding/ Aκουστική εξέταση Στο πρώτο μέρος της
Section 1: Listening and responding Presenter: Niki Farfara MGTAV VCE Seminar 7 August 2016 Section 1: Listening and responding Section 1: Listening and Responding/ Aκουστική εξέταση Στο πρώτο μέρος της
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
Advanced Subsidiary Unit 1: Understanding and Written Response
 Write your name here Surname Other names Edexcel GE entre Number andidate Number Greek dvanced Subsidiary Unit 1: Understanding and Written Response Thursday 16 May 2013 Morning Time: 2 hours 45 minutes
Write your name here Surname Other names Edexcel GE entre Number andidate Number Greek dvanced Subsidiary Unit 1: Understanding and Written Response Thursday 16 May 2013 Morning Time: 2 hours 45 minutes
Προγνωστικοί βιοδείκτες στην παιδική λευχαιμία
 27o Πανελλήνιο Συνέδριο Ελληνικής Εταιρίας Κοινωνικής Παιδιατρικής και Προαγωγής της Υγείας Προγνωστικοί βιοδείκτες στην παιδική λευχαιμία Παπαμικρούλης Γ.Α., Θωμόπουλος Θ., Πετρίδου Ε. Εργαστήριο Υγιεινής,
27o Πανελλήνιο Συνέδριο Ελληνικής Εταιρίας Κοινωνικής Παιδιατρικής και Προαγωγής της Υγείας Προγνωστικοί βιοδείκτες στην παιδική λευχαιμία Παπαμικρούλης Γ.Α., Θωμόπουλος Θ., Πετρίδου Ε. Εργαστήριο Υγιεινής,
± 20% ± 5% ± 10% RENCO ELECTRONICS, INC.
 RL15 RL16, RL17, RL18 MINIINDUCTORS CONFORMALLY COATED MARKING The nominal inductance is marked by a color code as listed in the table below. Color Black Brown Red Orange Yellow Green Blue Purple Grey
RL15 RL16, RL17, RL18 MINIINDUCTORS CONFORMALLY COATED MARKING The nominal inductance is marked by a color code as listed in the table below. Color Black Brown Red Orange Yellow Green Blue Purple Grey
5.4 The Poisson Distribution.
 The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1
 Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Προσομοίωση BP με το Bizagi Modeler
 Προσομοίωση BP με το Bizagi Modeler Α. Τσαλγατίδου - Γ.-Δ. Κάπος Πρόγραμμα Μεταπτυχιακών Σπουδών Τεχνολογία Διοίκησης Επιχειρησιακών Διαδικασιών 2017-2018 BPMN Simulation with Bizagi Modeler: 4 Levels
Προσομοίωση BP με το Bizagi Modeler Α. Τσαλγατίδου - Γ.-Δ. Κάπος Πρόγραμμα Μεταπτυχιακών Σπουδών Τεχνολογία Διοίκησης Επιχειρησιακών Διαδικασιών 2017-2018 BPMN Simulation with Bizagi Modeler: 4 Levels
ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE ΑΠΟ ΑΤΟΜΑ ΜΕ ΤΥΦΛΩΣΗ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions
 This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
Capacitors - Capacitance, Charge and Potential Difference
 Capacitors - Capacitance, Charge and Potential Difference Capacitors store electric charge. This ability to store electric charge is known as capacitance. A simple capacitor consists of 2 parallel metal
Capacitors - Capacitance, Charge and Potential Difference Capacitors store electric charge. This ability to store electric charge is known as capacitance. A simple capacitor consists of 2 parallel metal
ΓΕΩΜΕΣΡΙΚΗ ΣΕΚΜΗΡΙΩΗ ΣΟΤ ΙΕΡΟΤ ΝΑΟΤ ΣΟΤ ΣΙΜΙΟΤ ΣΑΤΡΟΤ ΣΟ ΠΕΛΕΝΔΡΙ ΣΗ ΚΤΠΡΟΤ ΜΕ ΕΦΑΡΜΟΓΗ ΑΤΣΟΜΑΣΟΠΟΙΗΜΕΝΟΤ ΤΣΗΜΑΣΟ ΨΗΦΙΑΚΗ ΦΩΣΟΓΡΑΜΜΕΣΡΙΑ
 ΕΘΝΙΚΟ ΜΕΣΟΒΙΟ ΠΟΛΤΣΕΧΝΕΙΟ ΣΜΗΜΑ ΑΓΡΟΝΟΜΩΝ-ΣΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΣΟΜΕΑ ΣΟΠΟΓΡΑΦΙΑ ΕΡΓΑΣΗΡΙΟ ΦΩΣΟΓΡΑΜΜΕΣΡΙΑ ΓΕΩΜΕΣΡΙΚΗ ΣΕΚΜΗΡΙΩΗ ΣΟΤ ΙΕΡΟΤ ΝΑΟΤ ΣΟΤ ΣΙΜΙΟΤ ΣΑΤΡΟΤ ΣΟ ΠΕΛΕΝΔΡΙ ΣΗ ΚΤΠΡΟΤ ΜΕ ΕΦΑΡΜΟΓΗ ΑΤΣΟΜΑΣΟΠΟΙΗΜΕΝΟΤ
ΕΘΝΙΚΟ ΜΕΣΟΒΙΟ ΠΟΛΤΣΕΧΝΕΙΟ ΣΜΗΜΑ ΑΓΡΟΝΟΜΩΝ-ΣΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΣΟΜΕΑ ΣΟΠΟΓΡΑΦΙΑ ΕΡΓΑΣΗΡΙΟ ΦΩΣΟΓΡΑΜΜΕΣΡΙΑ ΓΕΩΜΕΣΡΙΚΗ ΣΕΚΜΗΡΙΩΗ ΣΟΤ ΙΕΡΟΤ ΝΑΟΤ ΣΟΤ ΣΙΜΙΟΤ ΣΑΤΡΟΤ ΣΟ ΠΕΛΕΝΔΡΙ ΣΗ ΚΤΠΡΟΤ ΜΕ ΕΦΑΡΜΟΓΗ ΑΤΣΟΜΑΣΟΠΟΙΗΜΕΝΟΤ
Sampling Basics (1B) Young Won Lim 9/21/13
 Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Supporting Information
 Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ. Ψηφιακή Οικονομία. Διάλεξη 10η: Basics of Game Theory part 2 Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Ψηφιακή Οικονομία Διάλεξη 0η: Basics of Game Theory part 2 Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών Best Response Curves Used to solve for equilibria in games
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Ψηφιακή Οικονομία Διάλεξη 0η: Basics of Game Theory part 2 Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών Best Response Curves Used to solve for equilibria in games
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
The Simply Typed Lambda Calculus
 Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
NMBTC.COM /
 Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
SEN TRONIC AG 3-2 7 0 0 7 A 3 57 3 3 AB 93 :, C,! D 0 7 % 0 7 3 3 93 : 3 A 5 93 :
 # 3-270 07A35733 AB93:,C,!D 07% 0733 93: 3A593:!"#$%% &%&''()*%'+,-. &%&''(/*%'+0. 1*23 '4# 54/%6%7%53 *323 %7 77# %%3#% 8908/"/*55 :1$;/ = 7?@ > 7= 7 %! "$!"#$%&#%'(%%)*#$%&#%'(%#++#,-."/-0-1222"/-0-1
# 3-270 07A35733 AB93:,C,!D 07% 0733 93: 3A593:!"#$%% &%&''()*%'+,-. &%&''(/*%'+0. 1*23 '4# 54/%6%7%53 *323 %7 77# %%3#% 8908/"/*55 :1$;/ = 7?@ > 7= 7 %! "$!"#$%&#%'(%%)*#$%&#%'(%#++#,-."/-0-1222"/-0-1
Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo
 Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
2. THEORY OF EQUATIONS. PREVIOUS EAMCET Bits.
 EAMCET-. THEORY OF EQUATIONS PREVIOUS EAMCET Bits. Each of the roots of the equation x 6x + 6x 5= are increased by k so that the new transformed equation does not contain term. Then k =... - 4. - Sol.
EAMCET-. THEORY OF EQUATIONS PREVIOUS EAMCET Bits. Each of the roots of the equation x 6x + 6x 5= are increased by k so that the new transformed equation does not contain term. Then k =... - 4. - Sol.
EE101: Resonance in RLC circuits
 EE11: Resonance in RLC circuits M. B. Patil mbatil@ee.iitb.ac.in www.ee.iitb.ac.in/~sequel Deartment of Electrical Engineering Indian Institute of Technology Bombay I V R V L V C I = I m = R + jωl + 1/jωC
EE11: Resonance in RLC circuits M. B. Patil mbatil@ee.iitb.ac.in www.ee.iitb.ac.in/~sequel Deartment of Electrical Engineering Indian Institute of Technology Bombay I V R V L V C I = I m = R + jωl + 1/jωC
HOMEWORK 4 = G. In order to plot the stress versus the stretch we define a normalized stretch:
 HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
Figure 3 Three observations (Vp, Vs and density isosurfaces) intersecting in the PLF space. Solutions exist at the two indicated points.
 φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
Κάθε γνήσιο αντίγραφο φέρει υπογραφή του συγγραφέα. / Each genuine copy is signed by the author.
 Κάθε γνήσιο αντίγραφο φέρει υπογραφή του συγγραφέα. / Each genuine copy is signed by the author. 2012, Γεράσιμος Χρ. Σιάσος / Gerasimos Siasos, All rights reserved. Στοιχεία επικοινωνίας συγγραφέα / Author
Κάθε γνήσιο αντίγραφο φέρει υπογραφή του συγγραφέα. / Each genuine copy is signed by the author. 2012, Γεράσιμος Χρ. Σιάσος / Gerasimos Siasos, All rights reserved. Στοιχεία επικοινωνίας συγγραφέα / Author
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Approximation of distance between locations on earth given by latitude and longitude
 Approximation of distance between locations on earth given by latitude and longitude Jan Behrens 2012-12-31 In this paper we shall provide a method to approximate distances between two points on earth
Approximation of distance between locations on earth given by latitude and longitude Jan Behrens 2012-12-31 In this paper we shall provide a method to approximate distances between two points on earth
ΕΠΑΝΑΛΗΨΗ ΨΕΥΔΟΛΕΞΕΩΝ ΑΠΟ ΠΑΙΔΙΑ ΜΕ ΕΙΔΙΚΗ ΓΛΩΣΣΙΚΗ ΔΙΑΤΑΡΑΧΗ ΚΑΙ ΠΑΙΔΙΑ ΤΥΠΙΚΗΣ ΑΝΑΠΤΥΞΗΣ
 Σχολή Επιστημών Υγείας Πτυχιακή εργασία ΕΠΑΝΑΛΗΨΗ ΨΕΥΔΟΛΕΞΕΩΝ ΑΠΟ ΠΑΙΔΙΑ ΜΕ ΕΙΔΙΚΗ ΓΛΩΣΣΙΚΗ ΔΙΑΤΑΡΑΧΗ ΚΑΙ ΠΑΙΔΙΑ ΤΥΠΙΚΗΣ ΑΝΑΠΤΥΞΗΣ Άντρια Πολυκάρπου Λεμεσός, Μάιος 2017 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ
Σχολή Επιστημών Υγείας Πτυχιακή εργασία ΕΠΑΝΑΛΗΨΗ ΨΕΥΔΟΛΕΞΕΩΝ ΑΠΟ ΠΑΙΔΙΑ ΜΕ ΕΙΔΙΚΗ ΓΛΩΣΣΙΚΗ ΔΙΑΤΑΡΑΧΗ ΚΑΙ ΠΑΙΔΙΑ ΤΥΠΙΚΗΣ ΑΝΑΠΤΥΞΗΣ Άντρια Πολυκάρπου Λεμεσός, Μάιος 2017 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ
Δίκτυα Επικοινωνιών ΙΙ: OSPF Configuration
 Δίκτυα Επικοινωνιών ΙΙ: OSPF Configuration Δρ. Απόστολος Γκάμας Διδάσκων 407/80 gkamas@uop.gr Δίκτυα Επικοινωνιών ΙΙ Διαφάνεια 1 1 Dynamic Routing Configuration Router (config) # router protocol [ keyword
Δίκτυα Επικοινωνιών ΙΙ: OSPF Configuration Δρ. Απόστολος Γκάμας Διδάσκων 407/80 gkamas@uop.gr Δίκτυα Επικοινωνιών ΙΙ Διαφάνεια 1 1 Dynamic Routing Configuration Router (config) # router protocol [ keyword
6.1. Dirac Equation. Hamiltonian. Dirac Eq.
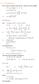 6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
Instruction Execution Times
 1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
ΙΔΡΥΜΑ. Θεσσαλονίκη, ύλα
 ΑΛΕΞΑΝΔΡΕΙΟ ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΑΣ ΓΕΩΠΟΝΙΑΣ, ΤΡΟΦΙΜΩΝ & ΔΙΑΤΡΟΦΗΔ ΗΣ ΤΜΗΜΑ ΔΙΑΤΡΟΦΗΣ & ΔΙΑΙΤΟΛΟΓΙΑΣ ΠΑΡΕΜΒΑΤΙΚΟ ΠΡΟΓΡΑΜΜΑ ΔΙΑΤΡΟΦΙΚΗΣ ΑΓΩΓΗΣ ΣΤΟΝ ΔΗΜΟ ΚΑΒΑΛΑΣ Πτυχιακή
ΑΛΕΞΑΝΔΡΕΙΟ ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΑΣ ΓΕΩΠΟΝΙΑΣ, ΤΡΟΦΙΜΩΝ & ΔΙΑΤΡΟΦΗΔ ΗΣ ΤΜΗΜΑ ΔΙΑΤΡΟΦΗΣ & ΔΙΑΙΤΟΛΟΓΙΑΣ ΠΑΡΕΜΒΑΤΙΚΟ ΠΡΟΓΡΑΜΜΑ ΔΙΑΤΡΟΦΙΚΗΣ ΑΓΩΓΗΣ ΣΤΟΝ ΔΗΜΟ ΚΑΒΑΛΑΣ Πτυχιακή
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
C.S. 430 Assignment 6, Sample Solutions
 C.S. 430 Assignment 6, Sample Solutions Paul Liu November 15, 2007 Note that these are sample solutions only; in many cases there were many acceptable answers. 1 Reynolds Problem 10.1 1.1 Normal-order
C.S. 430 Assignment 6, Sample Solutions Paul Liu November 15, 2007 Note that these are sample solutions only; in many cases there were many acceptable answers. 1 Reynolds Problem 10.1 1.1 Normal-order
*2354431106* GREEK 0543/02 Paper 2 Reading and Directed Writing May/June 2009
 UNIVERSITY OF CAMBRIDGE INTERNATIONAL EXAMINATIONS International General Certificate of Secondary Education *2354431106* GREEK 0543/02 Paper 2 Reading and Directed Writing May/June 2009 1 hour 30 minutes
UNIVERSITY OF CAMBRIDGE INTERNATIONAL EXAMINATIONS International General Certificate of Secondary Education *2354431106* GREEK 0543/02 Paper 2 Reading and Directed Writing May/June 2009 1 hour 30 minutes
Section 8.3 Trigonometric Equations
 99 Section 8. Trigonometric Equations Objective 1: Solve Equations Involving One Trigonometric Function. In this section and the next, we will exple how to solving equations involving trigonometric functions.
99 Section 8. Trigonometric Equations Objective 1: Solve Equations Involving One Trigonometric Function. In this section and the next, we will exple how to solving equations involving trigonometric functions.
Θέμα διπλωματικής εργασίας: «Από το «φρενοκομείο» στη Λέρο και την Ψυχιατρική Μεταρρύθμιση: νομικό πλαίσιο και ηθικοκοινωνικές διαστάσεις»
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΕΣ ΙΑΤΡΙΚΗΣ & ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΤΜΗΜΑΤΑ ΝΟΜΙΚΗΣ & ΘΕΟΛΟΓΙΑΣ ΔΙΑΤΜΗΜΑΤΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΣΥΓΧΡΟΝΕΣ ΙΑΤΡΙΚΕΣ ΠΡΑΞΕΙΣ: ΔΙΚΑΙΙΚΗ ΡΥΘΜΙΣΗ ΚΑΙ ΒΙΟΗΘΙΚΗ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΕΣ ΙΑΤΡΙΚΗΣ & ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΤΜΗΜΑΤΑ ΝΟΜΙΚΗΣ & ΘΕΟΛΟΓΙΑΣ ΔΙΑΤΜΗΜΑΤΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΣΥΓΧΡΟΝΕΣ ΙΑΤΡΙΚΕΣ ΠΡΑΞΕΙΣ: ΔΙΚΑΙΙΚΗ ΡΥΘΜΙΣΗ ΚΑΙ ΒΙΟΗΘΙΚΗ
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
2 Composition. Invertible Mappings
 Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
CE 530 Molecular Simulation
 C 53 olecular Siulation Lecture Histogra Reweighting ethods David. Kofke Departent of Cheical ngineering SUNY uffalo kofke@eng.buffalo.edu Histogra Reweighting ethod to cobine results taken at different
C 53 olecular Siulation Lecture Histogra Reweighting ethods David. Kofke Departent of Cheical ngineering SUNY uffalo kofke@eng.buffalo.edu Histogra Reweighting ethod to cobine results taken at different
TMA4115 Matematikk 3
 TMA4115 Matematikk 3 Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet Trondheim Spring 2010 Lecture 12: Mathematics Marvellous Matrices Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet
TMA4115 Matematikk 3 Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet Trondheim Spring 2010 Lecture 12: Mathematics Marvellous Matrices Andrew Stacey Norges Teknisk-Naturvitenskapelige Universitet
Ψηφιακή ανάπτυξη. Course Unit #1 : Κατανοώντας τις βασικές σύγχρονες ψηφιακές αρχές Thematic Unit #1 : Τεχνολογίες Web και CMS
 Ψηφιακή ανάπτυξη Course Unit #1 : Κατανοώντας τις βασικές σύγχρονες ψηφιακές αρχές Thematic Unit #1 : Τεχνολογίες Web και CMS Learning Objective : SEO και Analytics Fabio Calefato Department of Computer
Ψηφιακή ανάπτυξη Course Unit #1 : Κατανοώντας τις βασικές σύγχρονες ψηφιακές αρχές Thematic Unit #1 : Τεχνολογίες Web και CMS Learning Objective : SEO και Analytics Fabio Calefato Department of Computer
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Dr. D. Dinev, Department of Structural Mechanics, UACEG
 Lecture 4 Material behavior: Constitutive equations Field of the game Print version Lecture on Theory of lasticity and Plasticity of Dr. D. Dinev, Department of Structural Mechanics, UACG 4.1 Contents
Lecture 4 Material behavior: Constitutive equations Field of the game Print version Lecture on Theory of lasticity and Plasticity of Dr. D. Dinev, Department of Structural Mechanics, UACG 4.1 Contents
Calculating the propagation delay of coaxial cable
 Your source for quality GNSS Networking Solutions and Design Services! Page 1 of 5 Calculating the propagation delay of coaxial cable The delay of a cable or velocity factor is determined by the dielectric
Your source for quality GNSS Networking Solutions and Design Services! Page 1 of 5 Calculating the propagation delay of coaxial cable The delay of a cable or velocity factor is determined by the dielectric
ANSWERSHEET (TOPIC = DIFFERENTIAL CALCULUS) COLLECTION #2. h 0 h h 0 h h 0 ( ) g k = g 0 + g 1 + g g 2009 =?
 Teko Classes IITJEE/AIEEE Maths by SUHAAG SIR, Bhopal, Ph (0755) 3 00 000 www.tekoclasses.com ANSWERSHEET (TOPIC DIFFERENTIAL CALCULUS) COLLECTION # Question Type A.Single Correct Type Q. (A) Sol least
Teko Classes IITJEE/AIEEE Maths by SUHAAG SIR, Bhopal, Ph (0755) 3 00 000 www.tekoclasses.com ANSWERSHEET (TOPIC DIFFERENTIAL CALCULUS) COLLECTION # Question Type A.Single Correct Type Q. (A) Sol least
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Matrices and vectors. Matrix and vector. a 11 a 12 a 1n a 21 a 22 a 2n A = b 1 b 2. b m. R m n, b = = ( a ij. a m1 a m2 a mn. def
 Matrices and vectors Matrix and vector a 11 a 12 a 1n a 21 a 22 a 2n A = a m1 a m2 a mn def = ( a ij ) R m n, b = b 1 b 2 b m Rm Matrix and vectors in linear equations: example E 1 : x 1 + x 2 + 3x 4 =
Matrices and vectors Matrix and vector a 11 a 12 a 1n a 21 a 22 a 2n A = a m1 a m2 a mn def = ( a ij ) R m n, b = b 1 b 2 b m Rm Matrix and vectors in linear equations: example E 1 : x 1 + x 2 + 3x 4 =
Assalamu `alaikum wr. wb.
 LUMP SUM Assalamu `alaikum wr. wb. LUMP SUM Wassalamu alaikum wr. wb. Assalamu `alaikum wr. wb. LUMP SUM Wassalamu alaikum wr. wb. LUMP SUM Lump sum lump sum lump sum. lump sum fixed price lump sum lump
LUMP SUM Assalamu `alaikum wr. wb. LUMP SUM Wassalamu alaikum wr. wb. Assalamu `alaikum wr. wb. LUMP SUM Wassalamu alaikum wr. wb. LUMP SUM Lump sum lump sum lump sum. lump sum fixed price lump sum lump
Supplemental tables and figures
 Supplemental tables and figures Supplemental Table S1. Demographic and clinical parameters by quartiles of white Variables blood cell count. WBC quartiles( 1 9 cells/l) Q1(.6-5.3, n=2587) Q2(5.4-6.3, n=263)
Supplemental tables and figures Supplemental Table S1. Demographic and clinical parameters by quartiles of white Variables blood cell count. WBC quartiles( 1 9 cells/l) Q1(.6-5.3, n=2587) Q2(5.4-6.3, n=263)
 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation rate.3
.5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation rate.3
UNIVERSITY OF CAMBRIDGE INTERNATIONAL EXAMINATIONS International General Certificate of Secondary Education
 UNIVERSITY OF CAMBRIDGE INTERNATIONAL EXAMINATIONS International General Certificate of Secondary Education *2517291414* GREEK 0543/02 Paper 2 Reading and Directed Writing May/June 2013 1 hour 30 minutes
UNIVERSITY OF CAMBRIDGE INTERNATIONAL EXAMINATIONS International General Certificate of Secondary Education *2517291414* GREEK 0543/02 Paper 2 Reading and Directed Writing May/June 2013 1 hour 30 minutes
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΜΟΡΙΑΚΗ ΚΑΙ ΦΑΙΝΟΤΥΠΙΚΗ ΑΝΑΛΥΣΗ ΕΝΟΣ ΚΥΠΡΙΑΚΟΥ ΠΛΗΘΥΣΜΟΥ ΣΚΛΗΡΟΥ ΣΙΤΑΡΙΟΥ ΠΟΥ ΑΠΟΚΤΗΘΗΚΕ ΑΠΟ ΤΡΑΠΕΖΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΜΟΡΙΑΚΗ ΚΑΙ ΦΑΙΝΟΤΥΠΙΚΗ ΑΝΑΛΥΣΗ ΕΝΟΣ ΚΥΠΡΙΑΚΟΥ ΠΛΗΘΥΣΜΟΥ ΣΚΛΗΡΟΥ ΣΙΤΑΡΙΟΥ ΠΟΥ ΑΠΟΚΤΗΘΗΚΕ ΑΠΟ ΤΡΑΠΕΖΑ
Homework 8 Model Solution Section
 MATH 004 Homework Solution Homework 8 Model Solution Section 14.5 14.6. 14.5. Use the Chain Rule to find dz where z cosx + 4y), x 5t 4, y 1 t. dz dx + dy y sinx + 4y)0t + 4) sinx + 4y) 1t ) 0t + 4t ) sinx
MATH 004 Homework Solution Homework 8 Model Solution Section 14.5 14.6. 14.5. Use the Chain Rule to find dz where z cosx + 4y), x 5t 4, y 1 t. dz dx + dy y sinx + 4y)0t + 4) sinx + 4y) 1t ) 0t + 4t ) sinx
Η ΕΠΙΔΡΑΣΗ ΤΗΣ ΑΙΘΑΝΟΛΗΣ,ΤΗΣ ΜΕΘΑΝΟΛΗΣ ΚΑΙ ΤΟΥ ΑΙΘΥΛΟΤΡΙΤΟΤΑΓΗ ΒΟΥΤΥΛΑΙΘΕΡΑ ΣΤΙΣ ΙΔΙΟΤΗΤΕΣ ΤΗΣ ΒΕΝΖΙΝΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΚΑΒΑΛΑΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η ΕΠΙΔΡΑΣΗ ΤΗΣ ΑΙΘΑΝΟΛΗΣ,ΤΗΣ ΜΕΘΑΝΟΛΗΣ ΚΑΙ ΤΟΥ ΑΙΘΥΛΟΤΡΙΤΟΤΑΓΗ ΒΟΥΤΥΛΑΙΘΕΡΑ ΣΤΙΣ ΙΔΙΟΤΗΤΕΣ ΤΗΣ ΒΕΝΖΙΝΗΣ ΟΝΟΜΑΤΕΠΩΝΥΜΟ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΚΑΒΑΛΑΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η ΕΠΙΔΡΑΣΗ ΤΗΣ ΑΙΘΑΝΟΛΗΣ,ΤΗΣ ΜΕΘΑΝΟΛΗΣ ΚΑΙ ΤΟΥ ΑΙΘΥΛΟΤΡΙΤΟΤΑΓΗ ΒΟΥΤΥΛΑΙΘΕΡΑ ΣΤΙΣ ΙΔΙΟΤΗΤΕΣ ΤΗΣ ΒΕΝΖΙΝΗΣ ΟΝΟΜΑΤΕΠΩΝΥΜΟ
Inverse trigonometric functions & General Solution of Trigonometric Equations. ------------------ ----------------------------- -----------------
 Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια (MIS 365280)
 «Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
«Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
ΣΤΥΛΙΑΝΟΥ ΣΟΦΙΑ Socm09008@soc.aegean.gr
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΙΓΑΙΟΥ ΣΧΟΛΗ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΚΟΙΝΩΝΙΟΛΟΓΙΑΣ ΜΕΤΑΠΤΥΧΙΑΚΟ ΠΡΟΓΡΑΜΜΑ ΣΠΟΥΔΩΝ «ΕΡΕΥΝΑ ΓΙΑ ΤΗΝ ΤΟΠΙΚΗ ΚΟΙΝΩΝΙΚΗ ΑΝΑΠΤΥΞΗ ΚΑΙ ΣΥΝΟΧΗ» ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Θέμα: Διερεύνηση των απόψεων
ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΙΓΑΙΟΥ ΣΧΟΛΗ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΚΟΙΝΩΝΙΟΛΟΓΙΑΣ ΜΕΤΑΠΤΥΧΙΑΚΟ ΠΡΟΓΡΑΜΜΑ ΣΠΟΥΔΩΝ «ΕΡΕΥΝΑ ΓΙΑ ΤΗΝ ΤΟΠΙΚΗ ΚΟΙΝΩΝΙΚΗ ΑΝΑΠΤΥΞΗ ΚΑΙ ΣΥΝΟΧΗ» ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Θέμα: Διερεύνηση των απόψεων
