categorical Chris Parrish June 7, 2016
|
|
|
- Λαδων Ράγκος
- 6 χρόνια πριν
- Προβολές:
Transcript
1 categorical Chris Parrish June 7, 2016 Contents Waffle House 2 data exploratory data analysis Waffle Houses Marriage.s MedianAgeMarriage.s MedianAgeMarriage.s and Marriage.s model divorce rate ~ age at marriage map m model divorce rate ~ marriage rate map m model divorce rate ~ age at marriage and marriage rate map m model marriage rate ~ age at marriage map m plotting multivariate posteriors predictor residual plots counterfactual plots posterior prediction plots simulating spurious association milk 19 data map m5.5 with NA m5.5 complete cases m m legs 24 data exploratory data analysis model map parameter distributions samples parameter distributions milk 35 data exploratory data analysis model map parameter distributions plot parameter distribution
2 map plot parameter distribution map plot parameter distributions correlated predictor variables plants 46 data exploratory data analysis map parameter distributions plot parameter distributions map plot parameter distributions Dobe!Kung 52 data exploratory data analysis model map PI for male height reparametrize the model model map parameter distributions milk data map analysis unique intercepts lm 58 categorical references: - McElreath, Statistical Rethinking, chap 5, pp !Kung people - images of!kung people Waffle House library(rethinking) ## Loading required package: rstan ## Loading required package: ggplot2 ## rstan (Version , packaged: :54:41 UTC, GitRev: 05c3d0058b6a) ## For execution on a local, multicore CPU with excess RAM we recommend calling ## rstan_options(auto_write = TRUE) ## options(mc.cores = parallel::detectcores()) ## Loading required package: parallel 2
3 ## rethinking (Version 1.58) library(ggplot2) data ## R code 5.1 # load data data(waffledivorce) d <- WaffleDivorce str(d) ## 'data.frame': 50 obs. of 13 variables: ## $ Location : Factor w/ 50 levels "Alabama","Alaska",..: ## $ Loc : Factor w/ 50 levels "AK","AL","AR",..: ## $ Population : num ## $ MedianAgeMarriage: num ## $ Marriage : num ## $ Marriage.SE : num ## $ Divorce : num ## $ Divorce.SE : num ## $ WaffleHouses : int ## $ South : int ## $ Slaves1860 : int ## $ Population1860 : int ## $ PropSlaves1860 : num head(d) ## Location Loc Population MedianAgeMarriage Marriage Marriage.SE Divorce ## 1 Alabama AL ## 2 Alaska AK ## 3 Arizona AZ ## 4 Arkansas AR ## 5 California CA ## 6 Colorado CO ## Divorce.SE WaffleHouses South Slaves1860 Population1860 PropSlaves1860 ## ## ## ## ## ## exploratory data analysis Waffle Houses ggplot(d, aes(x = WaffleHouses / Population, y = Divorce)) + geom_point(shape = 20, color = "darkred") + geom_smooth(method = "lm") + labs(x = "Waffle Houses per million", y = "Divorce rate") 3
4 14 12 Divorce rate Waffle Houses per million Marriage.s Standardize marriage. d$marriage.s <- (d$marriage - mean(d$marriage))/sd(d$marriage) ggplot(d, aes(marriage.s, Divorce)) + geom_point(aes(x = Marriage.s, y = Divorce), shape = 20, color = "darkred") + geom_smooth(method = "lm") + labs(x = "Marriage.s", y = "Divorce") 4
5 12 Divorce Marriage.s M edianagem arriage.s Standardize median age at marriage. # standardize predictor d$medianagemarriage.s <- (d$medianagemarriage-mean(d$medianagemarriage))/ sd(d$medianagemarriage) ggplot(d, aes(medianagemarriage.s, Divorce)) + geom_point(aes(x = MedianAgeMarriage.s, y = Divorce), shape = 20, color = "darkred") + geom_smooth(method = "lm") + labs(x = "MedianAgeMarriage.s", y = "Divorce") 5
6 12 Divorce MedianAgeMarriage.s M edianagem arriage.s and M arriage.s How are marriage rate and median age at marriage related? ggplot(d, aes(medianagemarriage.s, Marriage.s)) + geom_point(shape = 20, color = "darkred") + geom_smooth(method = "lm") + labs(x = "MedianAgeMarriage.s", y = "Marriage.s") 6
7 3 2 1 Marriage.s MedianAgeMarriage.s model divorce rate ~ age at marriage. D i Normal(µ i, σ) µ i = α + β A A i α Normal(10, 10) β A Normal(0, 1) σ Uniform(0, 10) map m5.1 # fit model m5.1 <- map( alist( Divorce ~ dnorm( mu, sigma ), mu <- a + ba * MedianAgeMarriage.s, a ~ dnorm( 10, 10 ), ba ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data = d ) precis(m5.1) ## Mean StdDev 5.5% 94.5% ## a
8 ## ba ## sigma plot( precis(m5.1) ) a ba sigma Value ## R code 5.2 # compute percentile interval of mean MAM.seq <- seq( from=-3, to=3.5, length.out=30 ) mu <- link( m5.1, data=data.frame(medianagemarriage.s=mam.seq) ) ## [ 100 / 1000 ] [ 200 / 1000 ] [ 300 / 1000 ] [ 400 / 1000 ] [ 500 / 1000 ] [ 600 / 1000 ] [ 700 / 1000 ] [ 800 / 1000 ] [ 900 / 1000 ] [ 1000 / 1000 ] mu.pi <- apply( mu, 2, PI ) # plot it all plot( Divorce ~ MedianAgeMarriage.s, data=d, col=rangi2 ) abline( m5.1 ) ## Warning in abline(m5.1): only using the first two of 3 regression ## coefficients shade( mu.pi, MAM.seq ) 8
9 Divorce MedianAgeMarriage.s model divorce rate ~ marriage rate. D i Normal(µ i, σ) µ i = α + β R R i α Normal(10, 10) β R Normal(0, 1) σ Uniform(0, 10) map m5.2 ## R code 5.3 d$marriage.s <- (d$marriage - mean(d$marriage))/sd(d$marriage) m5.2 <- map( alist( Divorce ~ dnorm( mu, sigma ), mu <- a + br * Marriage.s, a ~ dnorm( 10, 10 ), br ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data = d ) precis(m5.2) ## Mean StdDev 5.5% 94.5% ## a ## br ## sigma plot( precis(m5.2) ) 9
10 a br sigma Value model divorce rate ~ age at marriage and marriage rate. D i Normal(µ i, σ) µ i = α + β R R i + β A A i α Normal(10, 10) β R Normal(0, 1) β A Normal(0, 1) σ Uniform(0, 10) map m5.3 ## R code 5.4 m5.3 <- map( alist( Divorce ~ dnorm( mu, sigma ), mu <- a + br*marriage.s + ba*medianagemarriage.s, a ~ dnorm( 10, 10 ), br ~ dnorm( 0, 1 ), ba ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data = d ) precis( m5.3 ) ## Mean StdDev 5.5% 94.5% ## a ## br ## ba ## sigma ## R code 5.5 plot( precis(m5.3) ) 10
11 a br ba sigma Value model marriage rate ~ age at marriage map m5.4 ## R code 5.6 m5.4 <- map( alist( Marriage.s ~ dnorm( mu, sigma ), mu <- a + b*medianagemarriage.s, a ~ dnorm( 0, 10 ), b ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data = d ) precis(m5.4) ## Mean StdDev 5.5% 94.5% ## a ## b ## sigma plot( precis(m5.4) ) 11
12 a b sigma Value plotting multivariate posteriors predictor residual plots ## R code 5.7 # compute expected value at MAP, for each State mu <- coef(m5.4)['a'] + coef(m5.4)['b']*d$medianagemarriage.s # compute residual for each State m.resid <- d$marriage.s - mu ## R code 5.8 plot( Marriage.s ~ MedianAgeMarriage.s, d, col=rangi2 ) abline( m5.4 ) ## Warning in abline(m5.4): only using the first two of 3 regression ## coefficients # loop over States for ( i in 1:length(m.resid) ) { x <- d$medianagemarriage.s[i] # x location of line segment y <- d$marriage.s[i] # observed endpoint of line segment # draw the line segment lines( c(x,x), c(mu[i],y), lwd=0.5, col=col.alpha("black",0.7) ) } 12
13 Marriage.s MedianAgeMarriage.s counterfactual plots ## R code 5.9 # prepare new counterfactual data A.avg <- mean( d$medianagemarriage.s ) R.seq <- seq( from=-3, to=3, length.out=30 ) pred.data <- data.frame( Marriage.s=R.seq, MedianAgeMarriage.s=A.avg ) # compute counterfactual mean divorce (mu) mu <- link( m5.3, data=pred.data ) ## [ 100 / 1000 ] [ 200 / 1000 ] [ 300 / 1000 ] [ 400 / 1000 ] [ 500 / 1000 ] [ 600 / 1000 ] [ 700 / 1000 ] [ 800 / 1000 ] [ 900 / 1000 ] [ 1000 / 1000 ] mu.mean <- apply( mu, 2, mean ) mu.pi <- apply( mu, 2, PI ) # simulate counterfactual divorce outcomes R.sim <- sim( m5.3, data=pred.data, n=1e4 ) ## [ 1000 / ] 13
14 [ 2000 / ] [ 3000 / ] [ 4000 / ] [ 5000 / ] [ 6000 / ] [ 7000 / ] [ 8000 / ] [ 9000 / ] [ / ] R.PI <- apply( R.sim, 2, PI ) # display predictions, hiding raw data with type="n" plot( Divorce ~ Marriage.s, data=d, type="n" ) mtext( "MedianAgeMarriage.s = 0" ) lines( R.seq, mu.mean ) shade( mu.pi, R.seq ) shade( R.PI, R.seq ) MedianAgeMarriage.s = 0 Divorce Marriage.s ## R code 5.10 R.avg <- mean( d$marriage.s ) A.seq <- seq( from=-3, to=3.5, length.out=30 ) pred.data2 <- data.frame( Marriage.s=R.avg, MedianAgeMarriage.s=A.seq ) mu <- link( m5.3, data=pred.data2 ) ## [ 100 / 1000 ] [ 200 / 1000 ] [ 300 / 1000 ] [ 400 / 1000 ] 14
15 [ 500 / 1000 ] [ 600 / 1000 ] [ 700 / 1000 ] [ 800 / 1000 ] [ 900 / 1000 ] [ 1000 / 1000 ] mu.mean <- apply( mu, 2, mean ) mu.pi <- apply( mu, 2, PI ) A.sim <- sim( m5.3, data=pred.data2, n=1e4 ) ## [ 1000 / ] [ 2000 / ] [ 3000 / ] [ 4000 / ] [ 5000 / ] [ 6000 / ] [ 7000 / ] [ 8000 / ] [ 9000 / ] [ / ] A.PI <- apply( A.sim, 2, PI ) plot( Divorce ~ MedianAgeMarriage.s, data=d, type="n" ) mtext( "Marriage.s = 0" ) lines( A.seq, mu.mean ) shade( mu.pi, A.seq ) shade( A.PI, A.seq ) Marriage.s = 0 Divorce MedianAgeMarriage.s 15
16 posterior prediction plots ## R code 5.11 # call link without specifying new data # so it uses original data mu <- link( m5.3 ) ## [ 100 / 1000 ] [ 200 / 1000 ] [ 300 / 1000 ] [ 400 / 1000 ] [ 500 / 1000 ] [ 600 / 1000 ] [ 700 / 1000 ] [ 800 / 1000 ] [ 900 / 1000 ] [ 1000 / 1000 ] # summarize samples across cases mu.mean <- apply( mu, 2, mean ) mu.pi <- apply( mu, 2, PI ) # simulate observations # again no new data, so uses original data divorce.sim <- sim( m5.3, n=1e4 ) ## [ 1000 / ] [ 2000 / ] [ 3000 / ] [ 4000 / ] [ 5000 / ] [ 6000 / ] [ 7000 / ] [ 8000 / ] [ 9000 / ] [ / ] divorce.pi <- apply( divorce.sim, 2, PI ) ## R code 5.12 plot( mu.mean ~ d$divorce, col=rangi2, ylim=range(mu.pi), xlab="observed divorce", ylab="predicted divorce" ) abline( a=0, b=1, lty=2 ) for ( i in 1:nrow(d) ) lines( rep(d$divorce[i],2), c(mu.pi[1,i],mu.pi[2,i]), col=rangi2 ) ## R code 5.13 identify( x=d$divorce, y=mu.mean, labels=d$loc, cex=0.8 ) 16
17 Predicted divorce ## integer(0) Observed divorce ## R code 5.14 # compute residuals divorce.resid <- d$divorce - mu.mean # get ordering by divorce rate o <- order(divorce.resid) # make the plot dotchart( divorce.resid[o], labels=d$loc[o], xlim=c(-6,5), cex=0.6 ) abline( v=0, col=col.alpha("black",0.2) ) for ( i in 1:nrow(d) ) { j <- o[i] # which State in order lines( d$divorce[j]-c(mu.pi[1,j],mu.pi[2,j]), rep(i,2) ) points( d$divorce[j]-c(divorce.pi[1,j],divorce.pi[2,j]), rep(i,2), pch=3, cex=0.6, col="gray" ) } 17
18 ME AR AL AK KY GA OK CO RI LA MS NH IN TN AZ SD OR VT WV NM WA MA MD KS IA OH DC NC DE MI HI TX VA MO WY IL MT FL CA NY PA WI SC NE CT UT ND MN NJ ID simulating spurious association ## R code 5.15 N <- 100 # number of cases x_real <- rnorm( N ) # x_real as Gaussian with mean 0 and stddev 1 x_spur <- rnorm( N, x_real ) # x_spur as Gaussian with mean=x_real y <- rnorm( N, x_real ) # y as Gaussian with mean=x_real d <- data.frame(y,x_real,x_spur) # bind all together in data frame pairs(~ y + x_real + x_spur, data=d, col="darkred") 18
19 y x_real x_spur demo.lm <- lm(y ~ x_real + x_spur, data=d) options(show.signif.stars=false) summary(demo.lm) ## ## Call: ## lm(formula = y ~ x_real + x_spur, data = d) ## ## Residuals: ## Min 1Q Median 3Q Max ## ## ## Coefficients: ## Estimate Std. Error t value Pr(> t ) ## (Intercept) ## x_real e-14 ## x_spur ## ## Residual standard error: on 97 degrees of freedom ## Multiple R-squared: , Adjusted R-squared: ## F-statistic: on 2 and 97 DF, p-value: 7.398e-16 milk library(rethinking) 19
20 data ## R code 5.16 data(milk) d <- milk str(d) ## 'data.frame': 29 obs. of 8 variables: ## $ clade : Factor w/ 4 levels "Ape","New World Monkey",..: ## $ species : Factor w/ 29 levels "A palliata","alouatta seniculus",..: ## $ kcal.per.g : num ## $ perc.fat : num ## $ perc.protein : num ## $ perc.lactose : num ## $ mass : num ## $ neocortex.perc: num 55.2 NA NA NA NA... map m5.5 with NA Problem here with incomplete cases. ## R code 5.17 # m5.5 <- map( # alist( # kcal.per.g ~ dnorm( mu, sigma ), # mu <- a + bn*neocortex.perc, # a ~ dnorm( 0, 100 ), # bn ~ dnorm( 0, 1 ), # sigma ~ dunif( 0, 1 ) # ), # data=d ) ## R code 5.18 d$neocortex.perc ## [1] NA NA NA NA NA ## [12] NA NA NA NA NA ## [23] NA NA m5.5 complete cases ## R code 5.19 dcc <- d[ complete.cases(d), ] a.start <- mean(dcc$kcal.per.g) bn.start <- 0 ## R code 5.20 m5.5 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), 20
21 mu <- a + bn*neocortex.perc, a ~ dnorm( 0, 100 ), bn ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 1 ) ), data = dcc, start = list(a=a.start, bn=bn.start)) ## R code 5.21 precis( m5.5, digits=3 ) ## Mean StdDev 5.5% 94.5% ## a ## bn ## sigma ## R code 5.22 coef(m5.5)["bn"] * ( ) ## bn ## ## R code 5.23 np.seq <- 0:100 pred.data <- data.frame( neocortex.perc=np.seq ) mu <- link( m5.5, data=pred.data, n=1e4 ) ## [ 1000 / ] [ 2000 / ] [ 3000 / ] [ 4000 / ] [ 5000 / ] [ 6000 / ] [ 7000 / ] [ 8000 / ] [ 9000 / ] [ / ] mu.mean <- apply( mu, 2, mean ) mu.pi <- apply( mu, 2, PI ) plot( kcal.per.g ~ neocortex.perc, data=dcc, col=rangi2 ) lines( np.seq, mu.mean ) lines( np.seq, mu.pi[1,], lty=2 ) lines( np.seq, mu.pi[2,], lty=2 ) 21
22 kcal.per.g ## R code 5.24 dcc$log.mass <- log(dcc$mass) neocortex.perc m5.6 ## R code 5.25 m5.6 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a + bm*log.mass, a ~ dnorm( 0, 100 ), bm ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 1 ) ), data=dcc ) precis(m5.6) ## Mean StdDev 5.5% 94.5% ## a ## bm ## sigma m5.7 Bad start value (1) ## R code 5.26 m5.7 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a + bn*neocortex.perc + bm*log.mass, 22
23 a ~ dnorm( 0, 100 ), bn ~ dnorm( 0, 1 ), bm ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 1 ) ), data=dcc ) precis(m5.7) ## Mean StdDev 5.5% 94.5% ## a ## bn ## bm ## sigma ## R code 5.27 mean.log.mass <- mean( log(dcc$mass) ) np.seq <- 0:100 pred.data <- data.frame( neocortex.perc=np.seq, log.mass=mean.log.mass ) mu <- link( m5.7, data=pred.data, n=1e4 ) ## [ 1000 / ] [ 2000 / ] [ 3000 / ] [ 4000 / ] [ 5000 / ] [ 6000 / ] [ 7000 / ] [ 8000 / ] [ 9000 / ] [ / ] mu.mean <- apply( mu, 2, mean ) mu.pi <- apply( mu, 2, PI ) plot( kcal.per.g ~ neocortex.perc, data=dcc, type="n" ) lines( np.seq, mu.mean ) lines( np.seq, mu.pi[1,], lty=2 ) lines( np.seq, mu.pi[2,], lty=2 ) 23
24 kcal.per.g neocortex.perc legs library(rethinking) library(ggplot2) library(cowplot) data Correlated predictors. ## R code 5.28 N <- 100 rho <- 0.7 x_pos <- rnorm( N ) x_neg <- rnorm( N, rho*x_pos, sqrt(1-rho^2) ) y <- rnorm( N, x_pos - x_neg ) d <- data.frame(y,x_pos,x_neg) Legs ## R code 5.29 N <- 100 height <- rnorm(n,10,2) leg_prop <- runif(n,0.4,0.5) leg_left <- leg_prop*height + rnorm( N, 0, 0.02 ) leg_right <- leg_prop*height + rnorm( N, 0, 0.02 ) # number of cases # correlation btw x_pos and x_neg # x_pos as Gaussian # x_neg correlated with x_pos # y equally associated with x_pos, x_neg # bind all together in data frame # number of individuals # sim total height of each # leg as proportion of height # sim left leg as proportion + error # sim right leg as proportion + error # combine into data frame 24
25 d <- data.frame(height,leg_left,leg_right) str(d) ## 'data.frame': 100 obs. of 3 variables: ## $ height : num ## $ leg_left : num ## $ leg_right: num exploratory data analysis ggplot(d, aes(leg_left, leg_right)) + geom_point(shape = 20, color = "darkred") + theme_gray() 6 5 leg_right leg_left ggplot(d, aes(leg_left, height)) + geom_point(shape = 20, color = "darkred") + geom_smooth() + theme_gray() 25
26 12.5 height leg_left model h i Normal(µ i, σ) µ i = α + β l leg l + β r leg r α Normal(10, 100) β l Normal(2, 10) β r Normal(2, 10) σ Uniform(0, 10) map Start values problem (4) ## R code 5.30 m5.8 <- map( alist( height ~ dnorm( mu, sigma ), mu <- a + bl*leg_left + br*leg_right, a ~ dnorm( 10, 100 ), bl ~ dnorm( 2, 10 ), br ~ dnorm( 2, 10 ), sigma ~ dunif( 0, 10 ) ), 26
27 data=d ) precis(m5.8) ## Mean StdDev 5.5% 94.5% ## a ## bl ## br ## sigma ## R code 5.31 plot(precis(m5.8)) a bl br sigma Value parameter distributions Plot parameter distributions # extract samples post <- extract.samples( m5.8 ) str(post) ## 'data.frame': obs. of 4 variables: ## $ a : num ## $ bl : num ## $ br : num ## $ sigma: num # plot each parameter distribution a.plot <- parameter.dist(parameter = "a", values = post$a) bl.plot <- parameter.dist(parameter = "bl", values = post$bl) 27
28 br.plot <- parameter.dist(parameter = "br", values = post$br) sigma.plot <- parameter.dist(parameter = "sigma", values = post$sigma) # display parameter distributions plot_grid(a.plot, bl.plot, br.plot, sigma.plot, labels=c("a", "bl", "br", "sigma"), ncol = 2, nrow = 2) 28
29 a 1.5 bl 0.20 density density a bl HPDI mean HPDI HPDI mean HPDI a bl br sigma density 0.10 density br sigma HPDI mean HPDI HPDI mean HPDI br sigma 29
30 samples ## R code 5.32 post <- extract.samples(m5.8) str(post) ## 'data.frame': obs. of 4 variables: ## $ a : num ## $ bl : num ## $ br : num ## $ sigma: num plot( bl ~ br, post, col=col.alpha(rangi2,0.1), pch=16 ) bl ggplot(post, aes(bl, br)) + geom_point(shape = 20, color = "darkred") + theme_gray() br 30
31 4 0 br bl ## R code 5.33 sum_blbr <- post$bl + post$br dens( sum_blbr, col=rangi2, lwd=2, xlab="sum of bl and br" ) 31
32 Density Start problem (2) sum of bl and br ## R code 5.34 m5.9 <- map( alist( height ~ dnorm( mu, sigma ), mu <- a + bl*leg_left, a ~ dnorm( 10, 100 ), bl ~ dnorm( 2, 10 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis(m5.9) ## Mean StdDev 5.5% 94.5% ## a ## bl ## sigma parameter distributions Plot parameter distributions # extract samples post <- extract.samples( m5.9 ) str(post) ## 'data.frame': obs. of 3 variables: ## $ a : num
33 ## $ bl : num ## $ sigma: num # plot each parameter distribution a.plot <- parameter.dist(parameter = "a", values = post$a) bl.plot <- parameter.dist(parameter = "bl", values = post$bl) sigma.plot <- parameter.dist(parameter = "sigma", values = post$sigma) # display parameter distributions plot_grid(a.plot, bl.plot, sigma.plot, labels=c("a", "bl", "sigma"), ncol = 2, nrow = 2) 33
34 a 1.5 bl 6 density 1.0 density a bl HPDI mean HPDI HPDI mean HPDI sigma a bl 7.5 density sigma HPDI mean HPDI sigma 34
35 milk library(rethinking) library(ggplot2) library(cowplot) theme_set(theme_gray()) data ## R code 5.35 data(milk) d <- milk str(d) ## 'data.frame': 29 obs. of 8 variables: ## $ clade : Factor w/ 4 levels "Ape","New World Monkey",..: ## $ species : Factor w/ 29 levels "A palliata","alouatta seniculus",..: ## $ kcal.per.g : num ## $ perc.fat : num ## $ perc.protein : num ## $ perc.lactose : num ## $ mass : num ## $ neocortex.perc: num 55.2 NA NA NA NA... exploratory data analysis Two predictor variables are associated. ggplot(d, aes(perc.fat, kcal.per.g)) + geom_point(shape = 20, color = "darkred") + geom_smooth() 35
36 kcal.per.g perc.fat ggplot(d, aes(perc.lactose, kcal.per.g)) + geom_point(shape = 20, color = "darkred") + geom_smooth() 36
37 kcal.per.g perc.lactose ggplot(d, aes(perc.fat, perc.lactose)) + geom_point(shape = 20, color = "darkred") + geom_smooth() 37
38 80 perc.lactose perc.fat model E i Normal(µ i, σ) µ i = α + β f perc.fat + β l perc.lactose α Normal(0.6, 10) β f Normal(0, 1) β l Normal(0, 1) σ Uniform(0, 10) map ## R code 5.36 # kcal.per.g regressed on perc.fat m5.10 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a + bf*perc.fat, a ~ dnorm( 0.6, 10 ), bf ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis( m5.10, digits=3 ) 38
39 ## Mean StdDev 5.5% 94.5% ## a ## bf ## sigma plot(precis( m5.10, digits=3 )) a bf sigma Value parameter distributions plot parameter distribution # extract samples post <- extract.samples( m5.10 ) str(post) ## 'data.frame': obs. of 3 variables: ## $ a : num ## $ bf : num ## $ sigma: num # plot parameter distribution bf.plot <- parameter.dist(parameter = "bf", values = post$bf) bf.plot 39
40 density bf HPDI mean HPDI bf map Start value (1) # kcal.per.g regressed on perc.lactose m5.11 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a + bl*perc.lactose, a ~ dnorm( 0.6, 10 ), bl ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis( m5.11, digits=3 ) ## Mean StdDev 5.5% 94.5% ## a ## bl ## sigma plot(precis( m5.11, digits=3 )) 40
41 a bl sigma Value plot parameter distribution # extract samples post <- extract.samples( m5.11 ) str(post) ## 'data.frame': obs. of 3 variables: ## $ a : num ## $ bl : num ## $ sigma: num # plot parameter distribution bl.plot <- parameter.dist(parameter = "bl", values = post$bl) bl.plot 41
42 density bl HPDI mean HPDI bl map ## R code 5.37 m5.12 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a + bf*perc.fat + bl*perc.lactose, a ~ dnorm( 0.6, 10 ), bf ~ dnorm( 0, 1 ), bl ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis( m5.12, digits=3 ) ## Mean StdDev 5.5% 94.5% ## a ## bf ## bl ## sigma plot(precis( m5.12, digits=3 )) 42
43 a bf bl sigma Value plot parameter distributions # extract samples post <- extract.samples( m5.12 ) str(post) ## 'data.frame': obs. of 4 variables: ## $ a : num ## $ bf : num ## $ bl : num ## $ sigma: num # plot each parameter distribution a.plot <- parameter.dist(parameter = "a", values = post$a) bf.plot <- parameter.dist(parameter = "bf", values = post$bf) bl.plot <- parameter.dist(parameter = "bl", values = post$bl) sigma.plot <- parameter.dist(parameter = "sigma", values = post$sigma) # display parameter distributions plot_grid(a.plot, bf.plot, bl.plot, sigma.plot, labels=c("a", "bf", "bl", "sigma"), ncol = 2, nrow = 2) 43
44 a 2.0 bf density 1.0 density a bf HPDI mean HPDI HPDI mean HPDI bl a bf sigma density 100 density bl sigma HPDI mean HPDI HPDI mean HPDI bl sigma 44
45 correlated predictor variables perc.f at and perc.lactose are inversely related ## R code 5.38 pairs( ~ kcal.per.g + perc.fat + perc.lactose, data=d, col=rangi2 ) kcal.per.g perc.fat perc.lactose ## R code 5.39 cor( d$perc.fat, d$perc.lactose ) ## [1] Simulation ## R code 5.40 data(milk) d <- milk sim.coll <- function( r=0.9 ) { d$x <- rnorm( nrow(d), mean=r*d$perc.fat, sd=sqrt( (1-r^2)*var(d$perc.fat) ) ) m <- lm( kcal.per.g ~ perc.fat + x, data=d ) sqrt( diag( vcov(m) ) )[2] # stddev of parameter } rep.sim.coll <- function( r=0.9, n=100 ) { stddev <- replicate( n, sim.coll(r) ) mean(stddev) } r.seq <- seq(from=0,to=0.99,by=0.01) stddev <- sapply( r.seq, function(z) rep.sim.coll(r=z,n=100) ) plot( stddev ~ r.seq, type="l", col=rangi2, lwd=2, xlab="correlation" ) 45
46 stddev correlation plants Post-treatment bias library(rethinking) library(ggplot2) library(cowplot) theme_set(theme_gray()) data ## R code 5.41 # number of plants N <- 100 # simulate initial heights h0 <- rnorm(n,10,2) # assign treatments and simulate fungus and growth treatment <- rep( 0:1, each=n/2 ) fungus <- rbinom( N, size=1, prob=0.5 - treatment*0.4 ) h1 <- h0 + rnorm(n, 5-3*fungus) # compose a clean data frame d <- data.frame( h0=h0, h1=h1, treatment=treatment, fungus=fungus ) 46
47 exploratory data analysis d2 <- d d2$treatment <- factor(d2$treatment) str(d2) ## 'data.frame': 100 obs. of 4 variables: ## $ h0 : num ## $ h1 : num ## $ treatment: Factor w/ 2 levels "0","1": ## $ fungus : int pairs(d2, col="darkred") h h1 treatment fungus ggplot(d2, aes(h0, h1, color = treatment)) + geom_point(shape = 20) + geom_smooth(method = "lm")
48 17.5 h treatment h0 map ## R code 5.42 m5.13 <- map( alist( h1 ~ dnorm(mu,sigma), mu <- a + bh*h0 + bt*treatment + bf*fungus, a ~ dnorm(0,100), c(bh,bt,bf) ~ dnorm(0,10), sigma ~ dunif(0,10) ), data=d, start = list(a = 0)) precis(m5.13) ## Mean StdDev 5.5% 94.5% ## a ## bh ## bt ## bf ## sigma
49 parameter distributions plot parameter distributions # extract samples post <- extract.samples( m5.13 ) str(post) ## 'data.frame': obs. of 5 variables: ## $ a : num ## $ bh : num ## $ bt : num ## $ bf : num ## $ sigma: num # plot each parameter distribution bh.plot <- parameter.dist(parameter = "bh", values = post$bh) bt.plot <- parameter.dist(parameter = "bt", values = post$bt) # display parameter distributions plot_grid(bh.plot, bt.plot, labels=c("bh", "bt"), ncol = 2, nrow = 1) bh 8 bt density 4 density bh bt HPDI mean HPDI HPDI mean HPDI bh bt map ## R code 5.43 m5.14 <- map( alist( h1 ~ dnorm(mu,sigma), 49
50 mu <- a + bh*h0 + bt*treatment, a ~ dnorm(0,100), c(bh,bt) ~ dnorm(0,10), sigma ~ dunif(0,10) ), data=d, start = list(a = 0)) precis(m5.14) ## Mean StdDev 5.5% 94.5% ## a ## bh ## bt ## sigma plot parameter distributions # extract samples post <- extract.samples( m5.14 ) str(post) ## 'data.frame': obs. of 4 variables: ## $ a : num ## $ bh : num ## $ bt : num ## $ sigma: num # plot each parameter distribution a.plot <- parameter.dist(parameter = "a", values = post$a) bh.plot <- parameter.dist(parameter = "bh", values = post$bh) bt.plot <- parameter.dist(parameter = "bt", values = post$bt) sigma.plot <- parameter.dist(parameter = "sigma", values = post$sigma) # display parameter distributions plot_grid(a.plot, bh.plot, bt.plot, sigma.plot, labels=c("a", "bh", "bt", "sigma"), ncol = 2, nrow = 2) 50
51 a 0.4 bh density 0.2 density a bh HPDI mean HPDI HPDI mean HPDI bt a sigma bh density 0.5 density bt sigma HPDI mean HPDI HPDI mean HPDI bt sigma 51
52 Dobe!Kung library(rethinking) library(ggplot2) library(cowplot) theme_set(theme_gray()) data ## R code 5.44 data(howell1) d <- Howell1 str(d) ## 'data.frame': 544 obs. of 4 variables: ## $ height: num ## $ weight: num ## $ age : num ## $ male : int exploratory data analysis d2 <- d d2$gender <- factor(d2$male, labels = c("female", "male")) str(d2) ## 'data.frame': 544 obs. of 5 variables: ## $ height: num ## $ weight: num ## $ age : num ## $ male : int ## $ gender: Factor w/ 2 levels "female","male": ggplot(d2, aes(weight, height, color = gender)) + geom_point(shape = 20) + scale_color_manual(values = c("skyblue", "sienna")) 52
53 150 height gender female male weight model h i Normal(µ i, σ) µ i = α + β m m i α Normal(178, 100) β m Normal(0, 10) σ Uniform(0, 50) map ## R code 5.45 m5.15 <- map( alist( height ~ dnorm( mu, sigma ), mu <- a + bm*male, a ~ dnorm( 178, 100 ), bm ~ dnorm( 0, 10 ), sigma ~ dunif( 0, 50 ) ), data=d, start = list(a = mean(d$height))) precis(m5.15) 53
54 ## Mean StdDev 5.5% 94.5% ## a ## bm ## sigma PI for male height ## R code 5.46 post <- extract.samples(m5.15) mu.male <- post$a + post$bm PI(mu.male) ## 5% 94% ## reparametrize the model model h i Normal(µ i, σ) µ i = β f f i + β m m i β f Normal(178, 100) β m Normal(178, 100) σ Uniform(0, 50) map ## R code 5.47 m5.15b <- map( alist( height ~ dnorm( mu, sigma ), mu <- af*(1-male) + am*male, af ~ dnorm( 178, 100 ), am ~ dnorm( 178, 100 ), sigma ~ dunif( 0, 50 ) ), data=d, start = list(af = mean(d$height), am = mean(d$height))) precis(m5.15b) ## Mean StdDev 5.5% 94.5% ## af ## am ## sigma
55 parameter distributions # extract samples post <- extract.samples( m5.15b ) str(post) ## 'data.frame': obs. of 3 variables: ## $ af : num ## $ am : num ## $ sigma: num # plot each parameter distribution af.plot <- parameter.dist(parameter = "af", values = post$af) am.plot <- parameter.dist(parameter = "am", values = post$am) # display parameter distributions plot_grid(af.plot, am.plot, labels=c("af", "am"), ncol = 2, nrow = 1) af 0.25 am density density af am HPDI mean HPDI HPDI mean HPDI af am milk data ## R code 5.48 data(milk) d <- milk unique(d$clade) ## [1] Strepsirrhine New World Monkey Old World Monkey Ape 55
56 ## Levels: Ape New World Monkey Old World Monkey Strepsirrhine ## R code 5.49 ( d$clade.nwm <- ifelse( d$clade=="new World Monkey", 1, 0 ) ) ## [1] ## R code 5.50 d$clade.owm <- ifelse( d$clade=="old World Monkey", 1, 0 ) d$clade.s <- ifelse( d$clade=="strepsirrhine", 1, 0 ) map ## R code 5.51 m5.16 <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a + b.nwm*clade.nwm + b.owm*clade.owm + b.s*clade.s, a ~ dnorm( 0.6, 10 ), b.nwm ~ dnorm( 0, 1 ), b.owm ~ dnorm( 0, 1 ), b.s ~ dnorm( 0, 1 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis(m5.16) ## Mean StdDev 5.5% 94.5% ## a ## b.nwm ## b.owm ## b.s ## sigma analysis ## R code 5.52 # sample posterior post <- extract.samples(m5.16) # compute averages for each category mu.ape <- post$a mu.nwm <- post$a + post$b.nwm mu.owm <- post$a + post$b.owm mu.s <- post$a + post$b.s # summarize using precis precis( data.frame(mu.ape,mu.nwm,mu.owm,mu.s) ) ## Mean StdDev ## mu.ape ## mu.nwm ## mu.owm
57 ## mu.s ## R code 5.53 diff.nwm.owm <- mu.nwm - mu.owm quantile( diff.nwm.owm, probs=c(0.025,0.5,0.975) ) ## 2.5% 50% 97.5% ## unique intercepts ## R code 5.54 ( d$clade_id <- coerce_index(d$clade) ) ## [1] ## R code 5.55 m5.16_alt <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a[clade_id], a[clade_id] ~ dnorm( 0.6, 10 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis( m5.16_alt, depth=2 ) ## Mean StdDev 5.5% 94.5% ## a[1] ## a[2] ## a[3] ## a[4] ## sigma ## R code 5.55 m5.16_alt <- map( alist( kcal.per.g ~ dnorm( mu, sigma ), mu <- a[clade_id], a[clade_id] ~ dnorm( 0.6, 10 ), sigma ~ dunif( 0, 10 ) ), data=d ) precis( m5.16_alt, depth=2 ) ## Mean StdDev 5.5% 94.5% ## a[1] ## a[2] ## a[3] ## a[4] ## sigma
58 lm # ## R code 5.56 # m5.17 <- lm( y ~ 1 + x, data=d ) # m5.18 <- lm( y ~ 1 + x + z + w, data=d ) # # ## R code 5.57 # m5.17 <- lm( y ~ 1 + x, data=d ) # m5.19 <- lm( y ~ x, data=d ) # # ## R code 5.58 # m5.20 <- lm( y ~ 0 + x, data=d ) # m5.21 <- lm( y ~ x - 1, data=d ) # # ## R code 5.59 # m5.22 <- lm( y ~ 1 + as.factor(season), data=d ) # # ## R code 5.60 # d$x2 <- d$x^2 # d$x3 <- d$x^3 # m5.23 <- lm( y ~ 1 + x + x2 + x3, data=d ) # # ## R code 5.61 # m5.24 <- lm( y ~ 1 + x + I(x^2) + I(x^3), data=d ) ## R code 5.62 data(cars) glimmer( dist ~ speed, data=cars ) ## alist( ## dist ~ dnorm( mu, sigma ), ## mu <- Intercept + ## b_speed*speed, ## Intercept ~ dnorm(0,10), ## b_speed ~ dnorm(0,10), ## sigma ~ dcauchy(0,2) ## ) 58
waffle Chris Parrish June 18, 2016
 waffle Chris Parrish June 18, 2016 Contents Waffle House 1 data..................................................... 2 exploratory data analysis......................................... 2 Waffle Houses.............................................
waffle Chris Parrish June 18, 2016 Contents Waffle House 1 data..................................................... 2 exploratory data analysis......................................... 2 Waffle Houses.............................................
m4.3 Chris Parrish June 16, 2016
 m4.3 Chris Parrish June 16, 2016 Contents!Kung model 1 data..................................................... 1 scatterplot with ggplot2....................................... 2 model....................................................
m4.3 Chris Parrish June 16, 2016 Contents!Kung model 1 data..................................................... 1 scatterplot with ggplot2....................................... 2 model....................................................
ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΙΓΑΙΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΤΗΣ ΙΟΙΚΗΣΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ ΜΕΤΑΠΤΥΧΙΑΚΟ ΙΠΛΩΜΑ ΙΟΙΚΗΣΗΣ ΕΠΙΧΕΙΡΗΣΕΩΝ ΜΕ. Ι..Ε.
 ΑΣΚΗΣΗ 1 ΟΜΑ Α 2 Στην ακόλουθη άσκηση σας δίνονται τα έξοδα ανά µαθητή και οι ετήσιοι µισθοί (κατά µέσο όρο) των δασκάλων για 51 πολιτείες της Αµερικής. Τα δεδοµένα είναι για τη χρονιά 1985. Οι µεταβλητές
ΑΣΚΗΣΗ 1 ΟΜΑ Α 2 Στην ακόλουθη άσκηση σας δίνονται τα έξοδα ανά µαθητή και οι ετήσιοι µισθοί (κατά µέσο όρο) των δασκάλων για 51 πολιτείες της Αµερικής. Τα δεδοµένα είναι για τη χρονιά 1985. Οι µεταβλητές
5.1 logistic regresssion Chris Parrish July 3, 2016
 5.1 logistic regresssion Chris Parrish July 3, 2016 Contents logistic regression model 1 1992 vote 1 data..................................................... 1 model....................................................
5.1 logistic regresssion Chris Parrish July 3, 2016 Contents logistic regression model 1 1992 vote 1 data..................................................... 1 model....................................................
United States of America
 page 1 ALABAMA AL Montgomery Most populous city: Birmingham 4,708,708 Square miles: 52,423 CST (UTC-6) EST (UTC-5) page 2 ALASKA AK Juneau Most populous city: Anchorage 698,473 Square miles: 656,425 AKST
page 1 ALABAMA AL Montgomery Most populous city: Birmingham 4,708,708 Square miles: 52,423 CST (UTC-6) EST (UTC-5) page 2 ALASKA AK Juneau Most populous city: Anchorage 698,473 Square miles: 656,425 AKST
519.22(07.07) 78 : ( ) /.. ; c (07.07) , , 2008
 .. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
.. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
SPRING /1-5/30 (PRE SPRING 12/1-5/30)
 SPRING 2019 1/1-5/30 (PRE SPRING 12/1-5/30) MISSY SIZES: XS - XXL PLUS SIZES: 1X - 3X ($5 UPCHARGE) p: 813-962-0900 f: 845-531-4814 e: CUSTOMERSERVICE@CAITEINC.COM START SHIP: 12/01 LKIC171 P94 DAHLIA
SPRING 2019 1/1-5/30 (PRE SPRING 12/1-5/30) MISSY SIZES: XS - XXL PLUS SIZES: 1X - 3X ($5 UPCHARGE) p: 813-962-0900 f: 845-531-4814 e: CUSTOMERSERVICE@CAITEINC.COM START SHIP: 12/01 LKIC171 P94 DAHLIA
Modern Regression HW #8 Solutions
 36-40 Modern Regression HW #8 Solutions Problem [25 points] (a) DUE: /0/207 at 3PM This is still a linear regression model the simplest possible one. That being the case, the solution we derived before
36-40 Modern Regression HW #8 Solutions Problem [25 points] (a) DUE: /0/207 at 3PM This is still a linear regression model the simplest possible one. That being the case, the solution we derived before
Ηλεκτρονικοί Υπολογιστές IV
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ Ηλεκτρονικοί Υπολογιστές IV Μοντέλα χρονολογικών σειρών Διδάσκων: Επίκουρος Καθηγητής Αθανάσιος Σταυρακούδης Άδειες Χρήσης Το παρόν εκπαιδευτικό υλικό
ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ Ηλεκτρονικοί Υπολογιστές IV Μοντέλα χρονολογικών σειρών Διδάσκων: Επίκουρος Καθηγητής Αθανάσιος Σταυρακούδης Άδειες Χρήσης Το παρόν εκπαιδευτικό υλικό
Wan Nor Arifin under the Creative Commons Attribution-ShareAlike 4.0 International License. 1 Introduction 1
 Linear Regression A Short Course on Data Analysis Using R Software (2017) Wan Nor Arifin (wnarifin@usm.my), Universiti Sains Malaysia Website: sites.google.com/site/wnarifin Wan Nor Arifin under the Creative
Linear Regression A Short Course on Data Analysis Using R Software (2017) Wan Nor Arifin (wnarifin@usm.my), Universiti Sains Malaysia Website: sites.google.com/site/wnarifin Wan Nor Arifin under the Creative
TABLES AND FORMULAS FOR MOORE Basic Practice of Statistics
 TABLES AND FORMULAS FOR MOORE Basic Practice of Statistics Exploring Data: Distributions Look for overall pattern (shape, center, spread) and deviations (outliers). Mean (use a calculator): x = x 1 + x
TABLES AND FORMULAS FOR MOORE Basic Practice of Statistics Exploring Data: Distributions Look for overall pattern (shape, center, spread) and deviations (outliers). Mean (use a calculator): x = x 1 + x
5.6 evaluating, checking, comparing Chris Parrish July 3, 2016
 5.6 evaluating, checking, comparing Chris Parrish July 3, 2016 Contents residuals 1 evaluating, checking, comparing 1 data..................................................... 1 model....................................................
5.6 evaluating, checking, comparing Chris Parrish July 3, 2016 Contents residuals 1 evaluating, checking, comparing 1 data..................................................... 1 model....................................................
Queensland University of Technology Transport Data Analysis and Modeling Methodologies
 Queensland University of Technology Transport Data Analysis and Modeling Methodologies Lab Session #7 Example 5.2 (with 3SLS Extensions) Seemingly Unrelated Regression Estimation and 3SLS A survey of 206
Queensland University of Technology Transport Data Analysis and Modeling Methodologies Lab Session #7 Example 5.2 (with 3SLS Extensions) Seemingly Unrelated Regression Estimation and 3SLS A survey of 206
Wan Nor Arifin under the Creative Commons Attribution-ShareAlike 4.0 International License. 1 Introduction 1
 Poisson Regression A Short Course on Data Analysis Using R Software (2017) Wan Nor Arifin (wnarifin@usm.my), Universiti Sains Malaysia Website: sites.google.com/site/wnarifin Wan Nor Arifin under the Creative
Poisson Regression A Short Course on Data Analysis Using R Software (2017) Wan Nor Arifin (wnarifin@usm.my), Universiti Sains Malaysia Website: sites.google.com/site/wnarifin Wan Nor Arifin under the Creative
Statistics 104: Quantitative Methods for Economics Formula and Theorem Review
 Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Biostatistics for Health Sciences Review Sheet
 Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
Οδηγίες χρήσης του R, μέρος 2 ο
 ΟδηγίεςχρήσηςτουR,μέρος2 ο Ελληνικά Ανπροσπαθήσουμεναγράψουμεελληνικάήναανοίξουμεκάποιοαρχείοδεδομένωνμε ελληνικούςχαρακτήρεςστοr,μπορείαντίγιαελληνικάναδούμελατινικούςχαρακτήρεςμε τόνουςήάλλακαλλικαντζαράκια.τότεδίνουμετηνπαρακάτωεντολήγιαναγυρίσειτοrστα
ΟδηγίεςχρήσηςτουR,μέρος2 ο Ελληνικά Ανπροσπαθήσουμεναγράψουμεελληνικάήναανοίξουμεκάποιοαρχείοδεδομένωνμε ελληνικούςχαρακτήρεςστοr,μπορείαντίγιαελληνικάναδούμελατινικούςχαρακτήρεςμε τόνουςήάλλακαλλικαντζαράκια.τότεδίνουμετηνπαρακάτωεντολήγιαναγυρίσειτοrστα
2018 Bowling.com Youth Open Championships OFFICIAL RESULTS Division: Doubles, U20 **All scholarship points are divided amongst team members** Rank
 2018 Bowling.com Youth Open Championships OFFICIAL RESULTS Division: Doubles, U20 **All scholarship points are divided amongst team members** Rank Team Name City State Series Total Points 1 Hodsdon/Olson
2018 Bowling.com Youth Open Championships OFFICIAL RESULTS Division: Doubles, U20 **All scholarship points are divided amongst team members** Rank Team Name City State Series Total Points 1 Hodsdon/Olson
DirichletReg: Dirichlet Regression for Compositional Data in R
 DirichletReg: Dirichlet Regression for Compositional Data in R Marco J. Maier Wirtschaftsuniversität Wien Abstract Full R Code for Maier, M. J. (2014). DirichletReg: Dirichlet Regression for Compositional
DirichletReg: Dirichlet Regression for Compositional Data in R Marco J. Maier Wirtschaftsuniversität Wien Abstract Full R Code for Maier, M. J. (2014). DirichletReg: Dirichlet Regression for Compositional
Aquinas College. Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET
 Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
FORMULAS FOR STATISTICS 1
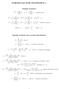 FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
Άσκηση 10, σελ. 119. Για τη μεταβλητή x (άτυπος όγκος) έχουμε: x censored_x 1 F 3 F 3 F 4 F 10 F 13 F 13 F 16 F 16 F 24 F 26 F 27 F 28 F
 Άσκηση 0, σελ. 9 από το βιβλίο «Μοντέλα Αξιοπιστίας και Επιβίωσης» της Χ. Καρώνη (i) Αρχικά, εισάγουμε τα δεδομένα στο minitab δημιουργώντας δύο μεταβλητές: τη x για τον άτυπο όγκο και την y για τον τυπικό
Άσκηση 0, σελ. 9 από το βιβλίο «Μοντέλα Αξιοπιστίας και Επιβίωσης» της Χ. Καρώνη (i) Αρχικά, εισάγουμε τα δεδομένα στο minitab δημιουργώντας δύο μεταβλητές: τη x για τον άτυπο όγκο και την y για τον τυπικό
Bayesian statistics. DS GA 1002 Probability and Statistics for Data Science.
 Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Start Random numbers Distributions p-value Confidence interval.
 Υπολογιστική Στατιστική με τη γλώσσα R Κατανομές και έλεγχοι υποθέσεων Αθανάσιος Σταυρακούδης http://stavrakoudis.econ.uoi.gr 19 Δεκεμβρίου 2013 1 / 33 Επισκόπηση 1 1 Start 2 Random numbers 3 Distributions
Υπολογιστική Στατιστική με τη γλώσσα R Κατανομές και έλεγχοι υποθέσεων Αθανάσιος Σταυρακούδης http://stavrakoudis.econ.uoi.gr 19 Δεκεμβρίου 2013 1 / 33 Επισκόπηση 1 1 Start 2 Random numbers 3 Distributions
Lampiran 1 Output SPSS MODEL I
 67 Variables Entered/Removed(b) Lampiran 1 Output SPSS MODEL I Model Variables Entered Variables Removed Method 1 CFO, ACCOTHER, ACCPAID, ACCDEPAMOR,. Enter ACCREC, ACCINV(a) a All requested variables
67 Variables Entered/Removed(b) Lampiran 1 Output SPSS MODEL I Model Variables Entered Variables Removed Method 1 CFO, ACCOTHER, ACCPAID, ACCDEPAMOR,. Enter ACCREC, ACCINV(a) a All requested variables
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Μηχανική Μάθηση Hypothesis Testing
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Μηχανική Μάθηση Hypothesis Testing Γιώργος Μπορμπουδάκης Τμήμα Επιστήμης Υπολογιστών Procedure 1. Form the null (H 0 ) and alternative (H 1 ) hypothesis 2. Consider
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Μηχανική Μάθηση Hypothesis Testing Γιώργος Μπορμπουδάκης Τμήμα Επιστήμης Υπολογιστών Procedure 1. Form the null (H 0 ) and alternative (H 1 ) hypothesis 2. Consider
[2] T.S.G. Peiris and R.O. Thattil, An Alternative Model to Estimate Solar Radiation
![[2] T.S.G. Peiris and R.O. Thattil, An Alternative Model to Estimate Solar Radiation [2] T.S.G. Peiris and R.O. Thattil, An Alternative Model to Estimate Solar Radiation](/thumbs/74/71129035.jpg) References [1] B.V.R. Punyawardena and Don Kulasiri, Stochastic Simulation of Solar Radiation from Sunshine Duration in Srilanka [2] T.S.G. Peiris and R.O. Thattil, An Alternative Model to Estimate Solar
References [1] B.V.R. Punyawardena and Don Kulasiri, Stochastic Simulation of Solar Radiation from Sunshine Duration in Srilanka [2] T.S.G. Peiris and R.O. Thattil, An Alternative Model to Estimate Solar
Solution Series 9. i=1 x i and i=1 x i.
 Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 651 APPENDIX B. BIBLIOGRAPHY 677 APPENDIX C. ANSWERS TO SELECTED EXERCISES 679
 APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
Table 1: Military Service: Models. Model 1 Model 2 Model 3 Model 4 Model 5 Model 6 Model 7 Model 8 Model 9 num unemployed mili mili num unemployed
 Tables: Military Service Table 1: Military Service: Models Model 1 Model 2 Model 3 Model 4 Model 5 Model 6 Model 7 Model 8 Model 9 num unemployed mili mili num unemployed mili 0.489-0.014-0.044-0.044-1.469-2.026-2.026
Tables: Military Service Table 1: Military Service: Models Model 1 Model 2 Model 3 Model 4 Model 5 Model 6 Model 7 Model 8 Model 9 num unemployed mili mili num unemployed mili 0.489-0.014-0.044-0.044-1.469-2.026-2.026
Generalized additive models in R
 www.nr.no Generalized additive models in R Magne Aldrin, Norwegian Computing Center and the University of Oslo Sharp workshop, Copenhagen, October 2012 Generalized Linear Models - GLM y Distributed with
www.nr.no Generalized additive models in R Magne Aldrin, Norwegian Computing Center and the University of Oslo Sharp workshop, Copenhagen, October 2012 Generalized Linear Models - GLM y Distributed with
 1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
Description of the PX-HC algorithm
 A Description of the PX-HC algorithm Let N = C c= N c and write C Nc K c= i= k= as, the Gibbs sampling algorithm at iteration m for continuous outcomes: Step A: For =,, J, draw θ m in the following steps:
A Description of the PX-HC algorithm Let N = C c= N c and write C Nc K c= i= k= as, the Gibbs sampling algorithm at iteration m for continuous outcomes: Step A: For =,, J, draw θ m in the following steps:
 η π 2 /3 χ 2 χ 2 t k Y 0/0, 0/1,..., 3/3 π 1, π 2,..., π k k k 1 β ij Y I i = 1,..., I p (X i = x i1,..., x ip ) Y i J (j = 1,..., J) x i Y i = j π j (x i ) x i π j (x i ) x (n 1 (x),..., n J (x))
η π 2 /3 χ 2 χ 2 t k Y 0/0, 0/1,..., 3/3 π 1, π 2,..., π k k k 1 β ij Y I i = 1,..., I p (X i = x i1,..., x ip ) Y i J (j = 1,..., J) x i Y i = j π j (x i ) x i π j (x i ) x (n 1 (x),..., n J (x))
A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads. Electronic Supplementary Material
 A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads Electronic Supplementary Material Ivan Jarić 1,2*, Jörn Gessner 1 and Mirjana Lenhardt 3 1 Leibniz-Institute
A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads Electronic Supplementary Material Ivan Jarić 1,2*, Jörn Gessner 1 and Mirjana Lenhardt 3 1 Leibniz-Institute
HISTOGRAMS AND PERCENTILES What is the 25 th percentile of a histogram? What is the 50 th percentile for the cigarette histogram?
 HISTOGRAMS AND PERCENTILES What is the 25 th percentile of a histogram? The point on the horizontal axis such that of the area under the histogram lies to the left of that point (and to the right) What
HISTOGRAMS AND PERCENTILES What is the 25 th percentile of a histogram? The point on the horizontal axis such that of the area under the histogram lies to the left of that point (and to the right) What
Γραµµική Παλινδρόµηση
 Κεφάλαιο 8 Γραµµική Παλινδρόµηση Η γραµµική παλινδρόµηση είναι ένα από τα πιο σηµαντικά ϑέµατα της Στατιστική ϑεωρείας. Στη συνέχεια αυτή η πολύ γνωστή µεθοδολογία ϑα αναπτυχθεί στην R µέσω των τύπων για
Κεφάλαιο 8 Γραµµική Παλινδρόµηση Η γραµµική παλινδρόµηση είναι ένα από τα πιο σηµαντικά ϑέµατα της Στατιστική ϑεωρείας. Στη συνέχεια αυτή η πολύ γνωστή µεθοδολογία ϑα αναπτυχθεί στην R µέσω των τύπων για
An Introduction to Splines
 An Introduction to Splines Trinity River Restoration Program Workshop on Outmigration: Population Estimation October 6 8, 2009 An Introduction to Splines 1 Linear Regression Simple Regression and the Least
An Introduction to Splines Trinity River Restoration Program Workshop on Outmigration: Population Estimation October 6 8, 2009 An Introduction to Splines 1 Linear Regression Simple Regression and the Least
10. ΠΟΛΛΑΠΛΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ
 0. ΠΟΛΛΑΠΛΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ 0. ΤΟ ΓΕΝΙΚΟ ΓΡΑΜΜΙΚΟ ΜΟΝΤΕΛΟ Συχνά στην πράξη το μοντέλο της απλής γραμμικής παλινδρόμησης είναι ανεπαρκές για την περιγραφή της μεταβλητότητας που υπάρχει στην εξαρτημένη
0. ΠΟΛΛΑΠΛΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ 0. ΤΟ ΓΕΝΙΚΟ ΓΡΑΜΜΙΚΟ ΜΟΝΤΕΛΟ Συχνά στην πράξη το μοντέλο της απλής γραμμικής παλινδρόμησης είναι ανεπαρκές για την περιγραφή της μεταβλητότητας που υπάρχει στην εξαρτημένη
ΠΟΛΛΑΠΛΗ ΠΑΛΙΝΔΡΟΜΗΣΗ: ΑΣΚΗΣΕΙΣ
 ΜΕΜ264: Εφαρμοσμένη Στατιστική 1 ΠΟΛΛΑΠΛΗ ΠΑΛΙΝΔΡΟΜΗΣΗ: ΑΣΚΗΣΕΙΣ 1. Σε μελέτη της επίδρασης γεωργικών χημικών στην προσρόφηση ιζημάτων και εδάφους, δίνονται στον πιο κάτω πίνακα 13 δεδομένα για το δείκτη
ΜΕΜ264: Εφαρμοσμένη Στατιστική 1 ΠΟΛΛΑΠΛΗ ΠΑΛΙΝΔΡΟΜΗΣΗ: ΑΣΚΗΣΕΙΣ 1. Σε μελέτη της επίδρασης γεωργικών χημικών στην προσρόφηση ιζημάτων και εδάφους, δίνονται στον πιο κάτω πίνακα 13 δεδομένα για το δείκτη
Bayesian Data Analysis, Midterm I
 Bayesian Data Analysis, Midterm I Bugra Gedik bgedik@cc.gatech.edu October 3, 4 Q1) I have used Gibs sampler to solve this problem. 5, iterations with burn-in value of 1, is used. The resulting histograms
Bayesian Data Analysis, Midterm I Bugra Gedik bgedik@cc.gatech.edu October 3, 4 Q1) I have used Gibs sampler to solve this problem. 5, iterations with burn-in value of 1, is used. The resulting histograms
ST5224: Advanced Statistical Theory II
 ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
8. ΑΠΛΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ Ι
 8. ΑΠΛΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ Ι Απλή γραμμική παλινδρόμηση είναι μία στατιστική μέθοδος που χρησιμοποιείται για τη μελέτη της σχέσης μεταξύ δύο ποσοτικών μεταβλητών εκ των οποίων μία είναι η ανεξάρτητη
8. ΑΠΛΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ Ι Απλή γραμμική παλινδρόμηση είναι μία στατιστική μέθοδος που χρησιμοποιείται για τη μελέτη της σχέσης μεταξύ δύο ποσοτικών μεταβλητών εκ των οποίων μία είναι η ανεξάρτητη
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
PENGARUHKEPEMIMPINANINSTRUKSIONAL KEPALASEKOLAHDAN MOTIVASI BERPRESTASI GURU TERHADAP KINERJA MENGAJAR GURU SD NEGERI DI KOTA SUKABUMI
 155 Lampiran 6 Yayan Sumaryana, 2014 PENGARUHKEPEMIMPINANINSTRUKSIONAL KEPALASEKOLAHDAN MOTIVASI BERPRESTASI GURU TERHADAP KINERJA MENGAJAR GURU SD NEGERI DI KOTA SUKABUMI Universitas Pendidikan Indonesia
155 Lampiran 6 Yayan Sumaryana, 2014 PENGARUHKEPEMIMPINANINSTRUKSIONAL KEPALASEKOLAHDAN MOTIVASI BERPRESTASI GURU TERHADAP KINERJA MENGAJAR GURU SD NEGERI DI KOTA SUKABUMI Universitas Pendidikan Indonesia
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
ΣΤΑΤΙΣΤΙΚΗ ΕΠΙΧΕΙΡΗΣΕΩΝ ΕΙΔΙΚΑ ΘΕΜΑΤΑ. Κεφάλαιο 13. Συμπεράσματα για τη σύγκριση δύο πληθυσμών
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΔΥΤΙΚΗΣ ΕΛΛΑΔΑΣ ΤΜΗΜΑ ΔΙΟΙΚΗΣΗΣ ΕΠΙΧΕΙΡΗΣΕΩΝ ΠΑΤΡΑΣ Εργαστήριο Λήψης Αποφάσεων & Επιχειρησιακού Προγραμματισμού Καθηγητής Ι. Μητρόπουλος ΣΤΑΤΙΣΤΙΚΗ ΕΠΙΧΕΙΡΗΣΕΩΝ ΕΙΔΙΚΑ ΘΕΜΑΤΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΔΥΤΙΚΗΣ ΕΛΛΑΔΑΣ ΤΜΗΜΑ ΔΙΟΙΚΗΣΗΣ ΕΠΙΧΕΙΡΗΣΕΩΝ ΠΑΤΡΑΣ Εργαστήριο Λήψης Αποφάσεων & Επιχειρησιακού Προγραμματισμού Καθηγητής Ι. Μητρόπουλος ΣΤΑΤΙΣΤΙΚΗ ΕΠΙΧΕΙΡΗΣΕΩΝ ΕΙΔΙΚΑ ΘΕΜΑΤΑ
Οδηγίες χρήσης του R, μέρος 1 ο. Κατεβάζουμε το λογισμικό από την ιστοσελίδα http://cran.cc.uoc.gr/bin/windows/base/
 ΟδηγίεςχρήσηςτουR,μέρος1 ο Προκαταρκτικά Κατεβάζουμετολογισμικόαπότηνιστοσελίδαhttp://cran.cc.uoc.gr/bin/windows/base/ Εγκαθιστούμετολογισμικόστονυπολογιστήμαςεκτελώνταςτοαρχείοπουκατεβάσαμε. ΤρέχουμετολογισμικόμεδιπλόκλικστομπλεεικονίδιοκαιβλέπουμετοπεριβάλλοντουR:
ΟδηγίεςχρήσηςτουR,μέρος1 ο Προκαταρκτικά Κατεβάζουμετολογισμικόαπότηνιστοσελίδαhttp://cran.cc.uoc.gr/bin/windows/base/ Εγκαθιστούμετολογισμικόστονυπολογιστήμαςεκτελώνταςτοαρχείοπουκατεβάσαμε. ΤρέχουμετολογισμικόμεδιπλόκλικστομπλεεικονίδιοκαιβλέπουμετοπεριβάλλοντουR:
3.4 SUM AND DIFFERENCE FORMULAS. NOTE: cos(α+β) cos α + cos β cos(α-β) cos α -cos β
 3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
( ) ( ) STAT 5031 Statistical Methods for Quality Improvement. Homework n = 8; x = 127 psi; σ = 2 psi (a) µ 0 = 125; α = 0.
 STAT 531 Statistical Methods for Quality Improvement Homework 3 4.8 n = 8; x = 17 psi; σ = psi (a) µ = 15; α =.5 Test H : µ = 15 vs. H 1 : µ > 15. Reject H if Z > Z α. x µ 17 15 Z = = =.88 σ n 8 Z α =
STAT 531 Statistical Methods for Quality Improvement Homework 3 4.8 n = 8; x = 17 psi; σ = psi (a) µ = 15; α =.5 Test H : µ = 15 vs. H 1 : µ > 15. Reject H if Z > Z α. x µ 17 15 Z = = =.88 σ n 8 Z α =
MATHACHij = γ00 + u0j + rij
 Stata output for Hierarchical Linear Models. ***************************************. * Unconditional Random Intercept Model. *************************************** MATHACHij = γ00 + u0j + rij. mixed
Stata output for Hierarchical Linear Models. ***************************************. * Unconditional Random Intercept Model. *************************************** MATHACHij = γ00 + u0j + rij. mixed
Web-based supplementary materials for Bayesian Quantile Regression for Ordinal Longitudinal Data
 Web-based supplementary materials for Bayesian Quantile Regression for Ordinal Longitudinal Data Rahim Alhamzawi, Haithem Taha Mohammad Ali Department of Statistics, College of Administration and Economics,
Web-based supplementary materials for Bayesian Quantile Regression for Ordinal Longitudinal Data Rahim Alhamzawi, Haithem Taha Mohammad Ali Department of Statistics, College of Administration and Economics,
Discontinuous Hermite Collocation and Diagonally Implicit RK3 for a Brain Tumour Invasion Model
 1 Discontinuous Hermite Collocation and Diagonally Implicit RK3 for a Brain Tumour Invasion Model John E. Athanasakis Applied Mathematics & Computers Laboratory Technical University of Crete Chania 73100,
1 Discontinuous Hermite Collocation and Diagonally Implicit RK3 for a Brain Tumour Invasion Model John E. Athanasakis Applied Mathematics & Computers Laboratory Technical University of Crete Chania 73100,
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Απλή Ευθύγραµµη Συµµεταβολή
 Απλή Ευθύγραµµη Συµµεταβολή Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές Τοµέας Φυτών Μεγάλης Καλλιέργειας και Οικολογίας, Εργαστήριο Γεωργίας Viola adorata Εισαγωγή Ανάλυση Παλινδρόµησης και Συσχέτιση Απλή
Απλή Ευθύγραµµη Συµµεταβολή Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές Τοµέας Φυτών Μεγάλης Καλλιέργειας και Οικολογίας, Εργαστήριο Γεωργίας Viola adorata Εισαγωγή Ανάλυση Παλινδρόµησης και Συσχέτιση Απλή
!"!"!!#" $ "# % #" & #" '##' #!( #")*(+&#!', & - #% '##' #( &2(!%#(345#" 6##7
 !"!"!!#" $ "# % #" & #" '##' #!( #")*(+&#!', '##' '# '## & - #% '##'.//0 #( 111111111111111111111111111111111111111111111111111 &2(!%#(345#" 6##7 11111111111111111111111111111111111111111111111111 11111111111111111111111111111111111111111111111111
!"!"!!#" $ "# % #" & #" '##' #!( #")*(+&#!', '##' '# '## & - #% '##'.//0 #( 111111111111111111111111111111111111111111111111111 &2(!%#(345#" 6##7 11111111111111111111111111111111111111111111111111 11111111111111111111111111111111111111111111111111
5.4 The Poisson Distribution.
 The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
SECTION II: PROBABILITY MODELS
 SECTION II: PROBABILITY MODELS 1 SECTION II: Aggregate Data. Fraction of births with low birth weight per province. Model A: OLS, using observations 1 260 Heteroskedasticity-robust standard errors, variant
SECTION II: PROBABILITY MODELS 1 SECTION II: Aggregate Data. Fraction of births with low birth weight per province. Model A: OLS, using observations 1 260 Heteroskedasticity-robust standard errors, variant
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
ΕΡΓΑΙΑ Εθηίκεζε αμίαο κεηαπώιεζεο ζπηηηώλ κε αλάιπζε δεδνκέλωλ. Παιεάο Δπζηξάηηνο
 ΕΡΓΑΙΑ Εθηίκεζε αμίαο κεηαπώιεζεο ζπηηηώλ κε αλάιπζε δεδνκέλωλ Παιεάο Δπζηξάηηνο ΑΘΗΝΑ 2014 1 ΠΔΡΙΔΥΟΜΔΝΑ 1) Δηζαγσγή 2) Πεξηγξαθηθή Αλάιπζε 3) ρέζεηο Μεηαβιεηώλ αλά 2 4) Πξνβιεπηηθά / Δξκελεπηηθά Μνληέια
ΕΡΓΑΙΑ Εθηίκεζε αμίαο κεηαπώιεζεο ζπηηηώλ κε αλάιπζε δεδνκέλωλ Παιεάο Δπζηξάηηνο ΑΘΗΝΑ 2014 1 ΠΔΡΙΔΥΟΜΔΝΑ 1) Δηζαγσγή 2) Πεξηγξαθηθή Αλάιπζε 3) ρέζεηο Μεηαβιεηώλ αλά 2 4) Πξνβιεπηηθά / Δξκελεπηηθά Μνληέια
CE 530 Molecular Simulation
 C 53 olecular Siulation Lecture Histogra Reweighting ethods David. Kofke Departent of Cheical ngineering SUNY uffalo kofke@eng.buffalo.edu Histogra Reweighting ethod to cobine results taken at different
C 53 olecular Siulation Lecture Histogra Reweighting ethods David. Kofke Departent of Cheical ngineering SUNY uffalo kofke@eng.buffalo.edu Histogra Reweighting ethod to cobine results taken at different
Ανάλυση Δεδομένων με χρήση του Στατιστικού Πακέτου R
 Ανάλυση Δεδομένων με χρήση του Στατιστικού Πακέτου R, Επίκουρος Καθηγητής, Τομέας Μαθηματικών, Σχολή Εφαρμοσμένων Μαθηματικών και Φυσικών Εφαρμογών, Εθνικό Μετσόβιο Πολυτεχνείο. Περιεχόμενα Εισαγωγή στη
Ανάλυση Δεδομένων με χρήση του Στατιστικού Πακέτου R, Επίκουρος Καθηγητής, Τομέας Μαθηματικών, Σχολή Εφαρμοσμένων Μαθηματικών και Φυσικών Εφαρμογών, Εθνικό Μετσόβιο Πολυτεχνείο. Περιεχόμενα Εισαγωγή στη
2. ΧΡΗΣΗ ΣΤΑΤΙΣΤΙΚΩΝ ΠΑΚΕΤΩΝ ΣΤΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ
 2. ΧΡΗΣΗ ΣΤΑΤΙΣΤΙΚΩΝ ΠΑΚΕΤΩΝ ΣΤΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ Η χρησιμοποίηση των τεχνικών της παλινδρόμησης για την επίλυση πρακτικών προβλημάτων έχει διευκολύνει εξαιρετικά από την χρήση διαφόρων στατιστικών
2. ΧΡΗΣΗ ΣΤΑΤΙΣΤΙΚΩΝ ΠΑΚΕΤΩΝ ΣΤΗ ΓΡΑΜΜΙΚΗ ΠΑΛΙΝΔΡΟΜΗΣΗ Η χρησιμοποίηση των τεχνικών της παλινδρόμησης για την επίλυση πρακτικών προβλημάτων έχει διευκολύνει εξαιρετικά από την χρήση διαφόρων στατιστικών
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ
 ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
Jesse Maassen and Mark Lundstrom Purdue University November 25, 2013
 Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
Optimizing Microwave-assisted Extraction Process for Paprika Red Pigments Using Response Surface Methodology
 2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
6.3 Forecasting ARMA processes
 122 CHAPTER 6. ARMA MODELS 6.3 Forecasting ARMA processes The purpose of forecasting is to predict future values of a TS based on the data collected to the present. In this section we will discuss a linear
122 CHAPTER 6. ARMA MODELS 6.3 Forecasting ARMA processes The purpose of forecasting is to predict future values of a TS based on the data collected to the present. In this section we will discuss a linear
Statistical Inference I Locally most powerful tests
 Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές. Εργαστήριο Γεωργίας. Viola adorata
 One-way ANOVA µε το SPSS Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές Τοµέας Φυτών Μεγάλης Καλλιέργειας και Οικολογίας, Εργαστήριο Γεωργίας Viola adorata To call in a statistician after the experiment is
One-way ANOVA µε το SPSS Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές Τοµέας Φυτών Μεγάλης Καλλιέργειας και Οικολογίας, Εργαστήριο Γεωργίας Viola adorata To call in a statistician after the experiment is
χ 2 test ανεξαρτησίας
 χ 2 test ανεξαρτησίας Καθηγητής Ι. Κ. ΔΗΜΗΤΡΙΟΥ demetri@econ.uoa.gr 7.2 Το χ 2 Τεστ Ανεξαρτησίας Tο χ 2 τεστ ανεξαρτησίας (όπως και η παλινδρόμηση) είναι στατιστικά εργαλεία για τον εντοπισμό σχέσεων μεταξύ
χ 2 test ανεξαρτησίας Καθηγητής Ι. Κ. ΔΗΜΗΤΡΙΟΥ demetri@econ.uoa.gr 7.2 Το χ 2 Τεστ Ανεξαρτησίας Tο χ 2 τεστ ανεξαρτησίας (όπως και η παλινδρόμηση) είναι στατιστικά εργαλεία για τον εντοπισμό σχέσεων μεταξύ
R & R- Studio. Πασχάλης Θρήσκος PhD Λάρισα
 R & R- Studio Πασχάλης Θρήσκος PhD Λάρισα 2016-2017 pthriskos@mnec.gr Εισαγωγή στο R Διαχείριση Δεδομένων R Project Περιγραφή του περιβάλλοντος του GNU προγράμματος R Project for Statistical Analysis Γραφήματα
R & R- Studio Πασχάλης Θρήσκος PhD Λάρισα 2016-2017 pthriskos@mnec.gr Εισαγωγή στο R Διαχείριση Δεδομένων R Project Περιγραφή του περιβάλλοντος του GNU προγράμματος R Project for Statistical Analysis Γραφήματα
 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation rate.3
.5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation rate.3
Αν οι προϋποθέσεις αυτές δεν ισχύουν, τότε ανατρέχουµε σε µη παραµετρικό τεστ.
 ΣΤ. ΑΝΑΛΥΣΗ ΙΑΣΠΟΡΑΣ (ANALYSIS OF VARIANCE - ANOVA) ΣΤ 1. Ανάλυση ιασποράς κατά µία κατεύθυνση. Όπως έχουµε δει στη παράγραφο Β 2, όταν θέλουµε να ελέγξουµε, αν η µέση τιµή µιας ποσοτικής µεταβλητής διαφέρει
ΣΤ. ΑΝΑΛΥΣΗ ΙΑΣΠΟΡΑΣ (ANALYSIS OF VARIANCE - ANOVA) ΣΤ 1. Ανάλυση ιασποράς κατά µία κατεύθυνση. Όπως έχουµε δει στη παράγραφο Β 2, όταν θέλουµε να ελέγξουµε, αν η µέση τιµή µιας ποσοτικής µεταβλητής διαφέρει
Parts Manual. Trio Mobile Surgery Platform. Model 1033
 Trio Mobile Surgery Platform Model 1033 Parts Manual For parts or technical assistance: Pour pièces de service ou assistance technique : Für Teile oder technische Unterstützung Anruf: Voor delen of technische
Trio Mobile Surgery Platform Model 1033 Parts Manual For parts or technical assistance: Pour pièces de service ou assistance technique : Für Teile oder technische Unterstützung Anruf: Voor delen of technische
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
DOUGLAS FIR BEETLE TRAP-SUPPRESSION STUDY STATISTICAL REPORT
 DOUGLAS FIR BEETLE TRAP-SUPPRESSION STUDY STATISTICAL REPORT Prepared for Dr. Robert Progar U.S. Forest Service Forest Sciences Laboratory Corvallis, Oregon January 2005 By Greg Brenner Pacific Analytics
DOUGLAS FIR BEETLE TRAP-SUPPRESSION STUDY STATISTICAL REPORT Prepared for Dr. Robert Progar U.S. Forest Service Forest Sciences Laboratory Corvallis, Oregon January 2005 By Greg Brenner Pacific Analytics
An Inventory of Continuous Distributions
 Appendi A An Inventory of Continuous Distributions A.1 Introduction The incomplete gamma function is given by Also, define Γ(α; ) = 1 with = G(α; ) = Z 0 Z 0 Z t α 1 e t dt, α > 0, >0 t α 1 e t dt, α >
Appendi A An Inventory of Continuous Distributions A.1 Introduction The incomplete gamma function is given by Also, define Γ(α; ) = 1 with = G(α; ) = Z 0 Z 0 Z t α 1 e t dt, α > 0, >0 t α 1 e t dt, α >
Στοιχεία από την r-project για την επεξεργασία και χαρτογράφηση χωρική κατανομή σημειακών παρατηρήσεων
 Στοιχεία από την r-project για την επεξεργασία και χαρτογράφηση χωρική κατανομή σημειακών παρατηρήσεων Ανάγνωση Δεδομένων # READ DATA # # εντοπισμός αρχείου filepath
Στοιχεία από την r-project για την επεξεργασία και χαρτογράφηση χωρική κατανομή σημειακών παρατηρήσεων Ανάγνωση Δεδομένων # READ DATA # # εντοπισμός αρχείου filepath
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
HOMEWORK 4 = G. In order to plot the stress versus the stretch we define a normalized stretch:
 HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
2. ΕΠΙΛΟΓΗ ΤΟΥ ΜΕΓΕΘΟΥΣ ΤΩΝ ΠΑΡΑΤΗΡΗΣΕΩΝ
 1. ΕΙΣΑΓΩΓΗ ΣΤΟ SPSS Το SPSS είναι ένα στατιστικό πρόγραμμα γενικής στατιστικής ανάλυσης αρκετά εύκολο στη λειτουργία του. Για να πραγματοποιηθεί ανάλυση χρονοσειρών με τη βοήθεια του SPSS θα πρέπει απαραίτητα
1. ΕΙΣΑΓΩΓΗ ΣΤΟ SPSS Το SPSS είναι ένα στατιστικό πρόγραμμα γενικής στατιστικής ανάλυσης αρκετά εύκολο στη λειτουργία του. Για να πραγματοποιηθεί ανάλυση χρονοσειρών με τη βοήθεια του SPSS θα πρέπει απαραίτητα
Estimation for ARMA Processes with Stable Noise. Matt Calder & Richard A. Davis Colorado State University
 Estimation for ARMA Processes with Stable Noise Matt Calder & Richard A. Davis Colorado State University rdavis@stat.colostate.edu 1 ARMA processes with stable noise Review of M-estimation Examples of
Estimation for ARMA Processes with Stable Noise Matt Calder & Richard A. Davis Colorado State University rdavis@stat.colostate.edu 1 ARMA processes with stable noise Review of M-estimation Examples of
Στατιστική Ανάλυση Δεδομένων II. Γραμμική Παλινδρόμηση με το S.P.S.S.
 Στατιστική Ανάλυση Δεδομένων II Γραμμική Παλινδρόμηση με το S.P.S.S. μέρος Α (απλή παλινδρόμηση) Νίκος Τσάντας Πρόγραμμα Μεταπτυχιακών Σπουδών Τμήμ. Μαθηματικών Μαθηματικά και Σύγχρονες Εφαρμογές Ακαδημαϊκό
Στατιστική Ανάλυση Δεδομένων II Γραμμική Παλινδρόμηση με το S.P.S.S. μέρος Α (απλή παλινδρόμηση) Νίκος Τσάντας Πρόγραμμα Μεταπτυχιακών Σπουδών Τμήμ. Μαθηματικών Μαθηματικά και Σύγχρονες Εφαρμογές Ακαδημαϊκό
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
1.1 t Rikon * --- Signif. codes: 0 *** ** 0.01 *
 Copyright (c) 004,005 Hidetoshi Shimodaira 1.. 1 1.1 t- 004-10-1 11:4:14 shimo Rikon 0.108355 0.04978.51 0.016136 * --- Signif. codes: 0 *** 0.001 ** 0.01 * 0.05. 0.1 1 Residual standard error: 0.03808
Copyright (c) 004,005 Hidetoshi Shimodaira 1.. 1 1.1 t- 004-10-1 11:4:14 shimo Rikon 0.108355 0.04978.51 0.016136 * --- Signif. codes: 0 *** 0.001 ** 0.01 * 0.05. 0.1 1 Residual standard error: 0.03808
SOLUTIONS TO MATH38181 EXTREME VALUES AND FINANCIAL RISK EXAM
 SOLUTIONS TO MATH38181 EXTREME VALUES AND FINANCIAL RISK EXAM Solutions to Question 1 a) The cumulative distribution function of T conditional on N n is Pr T t N n) Pr max X 1,..., X N ) t N n) Pr max
SOLUTIONS TO MATH38181 EXTREME VALUES AND FINANCIAL RISK EXAM Solutions to Question 1 a) The cumulative distribution function of T conditional on N n is Pr T t N n) Pr max X 1,..., X N ) t N n) Pr max
t ts P ALEPlot t t P rt P ts r P ts t r P ts
 t ts P ALEPlot 2 2 2 t t P rt P ts r P ts t r P ts t t r 1 t2 1 s r s r s r 1 1 tr s r t r s s rt t r s 2 s t t r r r t s s r t r t 2 t t r r t t2 t s s t t t s t t st 2 t t r t r t r s s t t r t s r t
t ts P ALEPlot 2 2 2 t t P rt P ts r P ts t r P ts t t r 1 t2 1 s r s r s r 1 1 tr s r t r s s rt t r s 2 s t t r r r t s s r t r t 2 t t r r t t2 t s s t t t s t t st 2 t t r t r t r s s t t r t s r t
TRIAXIAL TEST, CORPS OF ENGINEERS FORMAT
 TRIAXIAL TEST, CORPS OF ENGINEERS FORMAT .5 C, φ, deg Tan(φ) Total.7 2.2 Effective.98 8.33 Shear,.5.5.5 2 2.5 3 Total Normal, Effective Normal, Deviator,.5.25.75.5.25 2.5 5 7.5 Axial Strain, % Type of
TRIAXIAL TEST, CORPS OF ENGINEERS FORMAT .5 C, φ, deg Tan(φ) Total.7 2.2 Effective.98 8.33 Shear,.5.5.5 2 2.5 3 Total Normal, Effective Normal, Deviator,.5.25.75.5.25 2.5 5 7.5 Axial Strain, % Type of
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής
 Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Rod End > Ball Bearings Product Overview
 Product Overview Table of Contents EN4035 EN4036 FC...M / FCN...M ASNA2579E NSA8159 REP / RAP REP...F Page Il-3-4 Il-5-6 Il-7-8 Il-9-10 Il-11-12 Il-13-14 Il-15-16 EN4035 EN4035 L 06 P K A T Surface Treatment
Product Overview Table of Contents EN4035 EN4036 FC...M / FCN...M ASNA2579E NSA8159 REP / RAP REP...F Page Il-3-4 Il-5-6 Il-7-8 Il-9-10 Il-11-12 Il-13-14 Il-15-16 EN4035 EN4035 L 06 P K A T Surface Treatment
(i) Περιγραφική ανάλυση των μεταβλητών PRICE
 Με τις εντολές > data fdata names(fdata)=c("price", "SQFT",
Με τις εντολές > data fdata names(fdata)=c("price", "SQFT",
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ
 ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ Γηπισκαηηθή Δξγαζία ηνπ Φνηηεηή ηνπ ηκήκαηνο Ζιεθηξνιόγσλ Μεραληθώλ θαη Σερλνινγίαο Ζιεθηξνληθώλ
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ Γηπισκαηηθή Δξγαζία ηνπ Φνηηεηή ηνπ ηκήκαηνο Ζιεθηξνιόγσλ Μεραληθώλ θαη Σερλνινγίαο Ζιεθηξνληθώλ
Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov.
 A. ΈΛΕΓΧΟΣ ΚΑΝΟΝΙΚΟΤΗΤΑΣ A 1. Έλεγχος κανονικότητας Kolmogorov-Smirnov. Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov. Μηδενική υπόθεση:
A. ΈΛΕΓΧΟΣ ΚΑΝΟΝΙΚΟΤΗΤΑΣ A 1. Έλεγχος κανονικότητας Kolmogorov-Smirnov. Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov. Μηδενική υπόθεση:
Introduction to the ML Estimation of ARMA processes
 Introduction to the ML Estimation of ARMA processes Eduardo Rossi University of Pavia October 2013 Rossi ARMA Estimation Financial Econometrics - 2013 1 / 1 We consider the AR(p) model: Y t = c + φ 1 Y
Introduction to the ML Estimation of ARMA processes Eduardo Rossi University of Pavia October 2013 Rossi ARMA Estimation Financial Econometrics - 2013 1 / 1 We consider the AR(p) model: Y t = c + φ 1 Y
ΚΥΠΡΙΑΚΟΣ ΣΥΝΔΕΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY 21 ος ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ Δεύτερος Γύρος - 30 Μαρτίου 2011
 Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Το άτομο του Υδρογόνου
 Το άτομο του Υδρογόνου Δυναμικό Coulomb Εξίσωση Schrödinger h e (, r, ) (, r, ) E (, r, ) m ψ θφ r ψ θφ = ψ θφ Συνθήκες ψ(, r θφ, ) = πεπερασμένη ψ( r ) = 0 ψ(, r θφ, ) =ψ(, r θφ+, ) π Επιτρεπτές ενέργειες
Το άτομο του Υδρογόνου Δυναμικό Coulomb Εξίσωση Schrödinger h e (, r, ) (, r, ) E (, r, ) m ψ θφ r ψ θφ = ψ θφ Συνθήκες ψ(, r θφ, ) = πεπερασμένη ψ( r ) = 0 ψ(, r θφ, ) =ψ(, r θφ+, ) π Επιτρεπτές ενέργειες
1. Εισαγωγή...σελ Δεδομένα της εργασίας...σελ Μεθοδολογία...σελ Ανάλυση των δεδομένων.σελ Συγκριτικά αποτελέσματα..σελ.
 Περιεχόμενα 1. Εισαγωγή...σελ. 2 2. Δεδομένα της εργασίας......σελ. 3 3. Μεθοδολογία......σελ. 6 4. Ανάλυση των δεδομένων.σελ.13 5. Συγκριτικά αποτελέσματα..σελ.20 6. Συμπεράσματα.....σελ.20 7. Παράρτημα
Περιεχόμενα 1. Εισαγωγή...σελ. 2 2. Δεδομένα της εργασίας......σελ. 3 3. Μεθοδολογία......σελ. 6 4. Ανάλυση των δεδομένων.σελ.13 5. Συγκριτικά αποτελέσματα..σελ.20 6. Συμπεράσματα.....σελ.20 7. Παράρτημα
