Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia
|
|
|
- Ὀρέστης Ανδρεάδης
- 6 χρόνια πριν
- Προβολές:
Transcript
1 SUPPLEMENTAL MATERIAL FOR: Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia Matthew T. Witkowski, Luisa Cimmino, Yifang Hu, Thomas Trimarchi, Hiromi Tagoh, Mark D. McKenzie, Sarah A. Best, Laura Tuohey, Tracy A. Willson, Stephen L. Nutt, Meinrad Busslinger, Iannis Aifantis, Gordon K. Smyth, and Ross A. Dickins SUPPLEMENTAL METHODS Transgenic mice TRE-GFP-shRNA transgenes were detected by PCR using forward primers specific for each shrna (Ikaros.456: 5 -GTATTAATCTCTGAATACCACTGG; Luc.139: 5 - GTATTAATCAGAGACTTCAGGCGG) and a common reverse primer (GAAAGAACAATCAAGGGTCC) yielding a 21 bp product. The Vav-tTA transgene was detected tta specific forward (CCATACTCACTTTTGCCCTTTAG) and reverse (CAGCGCTGAGTGCATATAATGCA) primers, yielding a 221 bp product. Cell culture and western blotting OP9-DL1 stromal feeder cells were cultured in IMDM supplemented with 1% FBS, 1µg/mL streptomycin, 1U/mL penicillin, 1µM L-glutamine and 5µM β-mercaptoethanol and grown at 37 C in 1% CO 2. Wild type fetal liver cells were retrovirally co-infected with LMP-Cherrybased shrna vectors and MSCV-IRES-GFP vectors and cultured on an OP9-DL1 layer in media conditioned with 1ng/µL IL-7 (Peprotech) and 5ng/µL Flt3L for 12 days to induce T differentiation. 2Q T hybridoma cells were cultured similar to OP9-DL1 cells but with DME- KELSO replacing IMDM. Cell lysates were western blotted with anti-ikaros antibody E-2 (sc- 9861) (Santa Cruz Biotechnology, CA), anti-cleaved Notch1 (ICN1) antibody D3B8 (Cell Signaling, Boston, MA), anti-acetyl-histone H3 antibody (Millipore, Billerica, MA) and anti-actin antibody I-19 (sc-1616) (Santa Cruz Biotechnology). Leukemia transplantation Primary splenocytes from leukemic mice (predominantly leukemia cells) were transplanted by tail vein injection into immunocompromised Rag1 / recipient mice (2 x 1 6 cells/mouse). Transplant recipients generally developed overt signs of leukemia after approximately 2 weeks. For T-ALL transduction, 5 x 1 6 freshly harvested secondary leukemic spleen cells were spin-
2 infected at 1,2 rpm at 22 C onto a retrovirus-coated plate for 2 hours. Following centrifugation, cells were incubated in medium for 6 h at 37 C in 1% CO 2 and transplanted by tail vein injection into recipient mice. Doxycycline treatment was commenced upon detection of GFP + tumor cells in peripheral blood. Flow cytometry and blood analysis Mice were monitored for signs of leukemia (loss of activity or weight, breathing difficulty, enlarged lymph nodes, palpable splenomegaly). Single cell suspensions were prepared from bone marrow, thymus, and peripheral blood. Following red blood cell lysis, cells were stained with APC-conjugated anti-b22 (BD55392) or anti-cd8 (ebioscience ), or PEconjugated anti-cd4 (BD55373) or anti-mac1 (BD557397). Stained cells were analysed on a fluorescence activated cell sorter (BD FACSCalibur, BD Biosciences, San Jose, CA). To assess viability, cells were stained with AlexaFluor 647-conjugated AnnexinV (Invitrogen A2324) in AnnexinV binding buffer (BD556454). Stained cells were analysed on a fluorescence activated cell sorter (BD LSRII). For OP9-DL1 co-culture, cells were harvested and removed from the stromal layer for antibody staining. Biotinylated antibodies were used for lineage exclusion (Ter119-biotin Ly76, Gr1-biotin CR868C5, Mac1-biotin M1/7, B22-biotin RA36B2; generated in the WEHI Monoclonal Antibody Facility), then stained for CD8-PECy7 (Biolegend ), CD4-PE (WEHI GK1.5), CD44-APC (WEHI IM7.81) and CD25-PerCP.Cy5.5 (Biolegend PC621). Stained cells were analysed on a fluorescence activated cell sorter (BD LSRII). RNA-seq profiling of ALL65, ALL11, and ALL211 RNA samples were sequenced on an Illumina HiSeq 2 to produce 1 bp reads, either singleend (ALL65 and ALL11) or paired-end (ALL211). Reads were mapped to the mouse genome (mm1, NCBI build 38.1) by the subread aligner (7). Read counts were obtained for RefSeq Entrez genes using featurecounts (8). To cut down on variation not relevant to the current study, sex-linked genes (those on the Y chromosome plus Xist,), mitochondrial genes and unannotated genes without recognized gene symbols were excluded from the analysis. Remaining genes were filtered as not expressed if they failed to show at least one count per million reads in at least 3 samples. TMM normalization (9) was applied using edger software (1). The individual leukemias (ALL65, ALL11 and ALL211) were analyzed separately by computing predictive log2 fold changes between treated and untreated samples using edger with a prior count of 3. The predictive log folds are shrunk towards zero to avoid instability with small counts in one or more samples. Both technical and biological replicates of ALL11 were averaged for this purpose. Other statistical analyses were conducted using the limma software
3 package (11). Barcode plots for individual leukemias were ranked by predictive log fold changes and gene set tests used mean-rank enrichment tests (2). For analysis of biological replicates, read counts were transformed to log-counts-per-million and associated precision weights using the voom method (12). Statistical significance was assessed using linear models and empirical Bayes moderated t-statistics (13) controlling the false discovery rate at 5%. The analysis of the ALL11 biological replicates was a simple two group comparison. The combined analysis of all three leukemias was a paired analysis with the individual leukemias as blocks. For the combined analysis, the biological replicates of ALL11 were averaged by summing counts. Barcode plots of combined data were ranked by moderated t statistic (14) and gene set tests used rotation gene set testing (roast) (13, 15). Roast performs a hypothesis test that takes into account the directionality (up or down) and strength (log 2 fold change) of the genes in the set. Mutation analysis RNA-seq reads were searched for SNPs and other mutations using the exactsnp function of the subread package ( The ALL211 PEST domain sequence variant was identified by Sanger sequencing of a PCR product amplified from genomic DNA using forward (5 -CTACACAGCAGCCTCTCCAC) and reverse (5 - TGACCAGGAAAATCAAGGCTCT) primers. To assess whether the ALL11 Notch1 mutations were in cis or trans, approximately 2.4 kb of the coding region (spanning sequences encoding the HD and PEST domains) was PCR amplified from ALL11 cdna using forward (5 -GATCGGATCCATCAAGCGCTCTACAGTGG) and reverse (5 - GATCGTCGACGCAGAATGGTGTGCACAGGC) primers and cloned into pbluescript (Stratagene) using BamHI/SalI. The HD and PEST regions of several individual clones were sequenced using M13 forward (5 -TGTAAAACGACGGCCAGT) and reverse (5 - CAGGAAACAGCTATGAC) primers flanking the insert. Bio-ChIP-sequencing DP thymocytes (~2 x 1 8 ), which were enriched by CD8 MACS sorting from the thymus of Ikzf1 ihcd2/ihcd2 Rosa26 BirA/BirA mice, were used for chromatin precipitation by streptavidin pulldown (Bio-ChIP), as recently described in detail (16, 17). The precipitated genomic DNA was quantified by real-time PCR, and about 5 ng of precipitated DNA was used as starting material for generating single-end sequencing libraries selected for DNA fragments of 2 35 bp as described by Illumina s ChIP Sequencing sample preparation protocol. Sequencing was carried out using the Illumina/Solexa Genome Analyzer (GA) II systems according to manufacturer s guidelines. Sequence reads of 36 bp that passed the Illumina quality filtering were aligned against the mouse genome assembly version of July 27 (NCBI37/mm9) using the
4 Bowtie program version 12.5, allowing up to two mismatches and ignoring any read that would map more than once in the genome. Peaks were called by the MACS program (version ) and filtered for P values of < 1-1, as previously described (18). The peak-to-gene assignment was based on the RefSeq database, which was downloaded from UCSC on April 29th, 21. Peaks were assigned to genes in a stepwise manner by prioritizing genes containing peaks in their promoter and/or gene body, as described (18). This procedure essentially assigned peaks to a gene if they were located in a region spanning from 5 kb upstream of the TSS to 5 kb downstream of the TES of a given gene. Primary human primary samples Primary T-ALL samples were provided by the Childrens Oncology Group with informed consent and analyzed under the supervision of the New York University Langone Medical Center Institutional Review Board. Total RNA was extracted from samples using the RNeasy Plus mini kit (Life Technologies, Carlsbad, CA). Samples were subject to rrna removal using the Ribo- Zero kit (Epicentre, Madison, WI) according to manufacturer instructions. The resulting RNA samples were then used as input for library construction using the dutp method as described (19). RNA libraries were sequenced on Illumina HiSeq 2 and 25 machines to produce 5bp paired-end reads. Reads were aligned to the human hg19 genome using TopHat (2) v1.4 with default parameters. FPKM (fragments per kilobase per million reads) expression values were generated by Cufflinks (21) v1.3 using RefSeq gene annotation. Genes with reduced expression in T-ALL relative to thymocytes were ranked by Students s t test P value following removal of genes that had zero FKPM values in at least one sample. SUPPLEMENTAL REFERENCES 1. Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & Development 26 Aug 1; 2(15): Michaud J, Simpson KM, Escher R, Buchet-Poyau K, Beissbarth T, Carmichael C, et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics 28; 9: Zeller KI, Zhao X, Lee CWH, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA 26 Nov 21; 13(47): Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, et al. The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 213 Jun; 14(6): Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 214 Oct; 2(1):
5 6. Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci U S A 214 Nov 18; 111(46): E Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 213 May 1; 41(1): e Liao Y, Smyth GK, Shi W. featurecounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 213 Nov Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 21 Jan 1; 11(3): R Robinson MD, McCarthy DJ, Smyth GK. edger: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 21 Jan 1; 26(1): Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 215; 43: in press. 12. Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 214; 15(2): R Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 24 Jan 1; 3: Article Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature Medicine 29 Aug 1; 15(8): Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 21 Sep 1; 26(17): Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, et al. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-b cells. Immunity 211 Feb 25; 34(2): Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, et al. Stagespecific control of early B cell development by the transcription factor Ikaros. Nat Immunol 214; 15(3): Revilla-I-Domingo R, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 212; 31(14): Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 29 Oct; 37(18): e Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 29 May 1; 25(9): Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 21 May; 28(5):
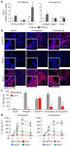
6 Figure S1 A ATG STOP Ikaros.279 Ikaros.456 mrna isoform Ik1 Exon CDS B 1 C survival (%) D Ren.713 (n=1) 2 Ikaros.279 (n=6) Ikaros.456 (n=7) age (days) 4 2 spleen thymus Ren.713 Ikaros.456 cell count CD GFP CD8
7 Figure S2 A X Vav-tTA B Vav-tTA; TRE-GFPshIkaros leukemia transplant 3 mice untreated 3 mice Dox 3d FACS sort GFP+ leukemia cells VAV-tTA; TRE-GFP-shIkaros untreated + Dox shrna on: Ikaros knockdown shrna off: Ikaros restoration
8 Ikzf1 Fignl1 untreated 3 days Dox B A Figure S GFP Freq untreated d3 Dox C ALL11: RNA Seq Average log expression log fold change Ikzf1 398 genes 2943 genes 2-6 log2 fold change average expression (log2 counts per million) ALL11: RNA-Seq
9 Figure S4 A 4.7 Enrichment ALL65 RNA-Seq Weng Notch withdrawal genes Mean-rank gene set test Red bars: genes increased upon Notch withdrawal B 4.5 Enrichment ALL11 RNA-Seq Weng Notch withdrawal genes Mean-rank gene set test Red bars: genes increased upon Notch withdrawal P<1-5 P<1-4 increased upon Ikaros restoration decreased upon Ikaros restoration increased upon Ikaros restoration decreased upon Ikaros restoration P<1-5 P<1-8 Enrichment Enrichment 2.5 Blue bars: genes decreased upon Notch withdrawal 3 Blue bars: genes decreased upon Notch withdrawal C 3.6 Enrichment ALL211 RNA-Seq Weng Notch withdrawal genes Mean-rank gene set test Red bars: genes increased upon Notch withdrawal P<1-6 increased upon Ikaros restoration decreased upon Ikaros restoration P<1-11 Enrichment 4.1 Blue bars: genes decreased upon Notch withdrawal
10 Figure S5 A Transplant ut Disease onset Dox d3 d7 Dox Harvest recipients -7d -3d ut C Percent survival 1 5 ALL211 untreated negative shikaros Dox ICN1 ALL65 ALL11 ALL211 D Percent survival Days post dox ALL11 untreated negative shikaros ICN1 Dox B Percent survival 1 5 ALL65 untreated negative shikaros ICN1 Dox E 5 kda ALL65 d Days post dox d3 ALL11 d d3 Ik1 Ik Days post dox 37 kda Actin
11 Figure S6 A 5 kda 37 kda relapse d29 Dox d d3 #1 #2 Ik1 * Ik2 * B ALL65 Relative expression to HPRT Ikaros Exons 3-4 Ikaros Exons 7-8 Hes1 Relative expression to HPRT d d3 Relapse d d3 Relapse Relative expression to Gapdh d d3 Relapse 37 kda Actin ALL211 Relative expression to HPRT d d3 Relapse Relative expression to HPRT d d3 Relapse Relative expression to Gapdh d d3 Relapse ALL11 Relative expression to HPRT d d3 Relapse Relative expression to HPRT d d3 Relapse Relative expression to Gapdh d d3 Relapse
12 Notch1 chr2: (Notch1 Figure S7 A Notch1 chr2: (Notch1 Exon 26) Notch1 chr2: Notch1 Exon 34) ALL211 ALL ALL65 (P41S) ALL65 (N2385T) ALL11 ALL11 Sequence Sequence B Notch1 chr2: (Notch1 Exon 27) Notch1 Notch1 chr2: (Notch1 Exon 34) Notch ALL211 ALL ALL65 ALL ALL11 (L1668P) ALL11 (S2398X) Sequence Sequence Notch1 Notch1 C Notch1 chr2: (Notch1 Exon 27) D ALL211 genomic DNA Notch1 Sanger Sequencing 46 PEST DOMAIN ALL11 (wildtype) ALL211 (Y176S) 65 P S S M V C G C A ALL211 (S2446fsX1) ALL E HD DOMAIN P S S X ALL11 cdna Notch1 Sanger Sequencing M V PEST DOMAIN ALL11 Sequence Exon 27 L1668P 68P (HD domain) Clone #1 wildtype Clone #2 c.52t>c (L1668P) Clone #1 c.7192c>a (S2398X) V Y L E I L Clone #2 wildtype V Y P E I L V S X A A L V S S A A L Notch1
13 Figure S8 A Combined T-ALL Zeller Myc-regulated genes: NEGATIVE correlation (P=.547) C Reads Enrichment increased upon Ikaros restoration in leukemia Red bars: genes induced directly by Myc decreased upon Ikaros restoration in leukemia D Reads 1 Myc 5 kb Enrichment 2.6 Blue bars: genes repressed directly by Myc Myc Pvt1 1 kb N-Me B 3.3 Combined T-ALL Myc cluster (Mingueneau) Enrichment (P=.547) Enrichment increased upon Ikaros restoration in leukemia decreased upon Ikaros restoration in leukemia Black bars: genes where expression correlates with Myc expression throught thymocyte development
14 Figure S9 CUTLL1 HPB-ALL GSI (hours) IKZF1 -ACTIN
15 SUPPLEMENTAL FIGURE LEGENDS Figure S1. (A) Schematic of murine Ikzf1/Ikaros full-length Ik1 isoform, showing sites targeted by the Ikaros.279 and Ikaros.456 shrnas. (B) Kaplan-Meier survival curves of recipient mice reconstituted with fetal liver hematopoietic stem progenitor cells infected with LMP vectors expressing Ikaros shrnas or Ren.713 as indicated. (C) Enlarged thymus and spleen of a representative leukemic Ikaros.456 mouse compared with a healthy Ren.713 control. (D) Representative flow cytometry of splenocytes isolated from a leukemic Ikaros.456 mouse showing accumulation of GFP + CD4 + CD8 + cells. Figure S2. (A) Mating strategy to generate a mouse model of inducible Ikaros restoration in T- ALL. (B) Strategy for tet-regulated Ikaros restoration in T-ALL in vivo. Figure S3. (A) Histogram of GFP expression in cell suspensions from spleens of leukemic recipient mice following transplant with Vav-tTA;TRE-GFP-shIkaros leukemia ALL11. Mouse treatments and gates for sorting of GFP + leukemia cells by FACS are indicated. (B) Scatter plot of RNA-seq differential expression upon Ikaros restoration in ALL11 (comparing 3 days Dox with untreated, averaged from 3 mice per condition). Genes with significant expression changes (FDR<.5) are highlighted, and Ikaros is indicated. (C) ALL11 RNA-seq tracks for Ikzf1/Ikaros and neighbouring control gene Fignl1 for triplicate untreated and Dox treated mice. Figure S4. Gene set analysis barcode plots of RNA-seq differential gene expression upon Ikaros restoration in ALL65 (upper), ALL11 (middle), and ALL211 (lower), with genes horizontally ranked by predictive log2 fold changes. Genes upregulated upon Ikaros restoration are shaded pink (logfc > 1.2) and downregulated genes are shaded blue (logfc < 1.2). As for Figure 2E, overlaid are a previously described set of genes induced (red bars) or repressed (blue bars) upon Notch inhibition in a murine T-ALL cell line (1). Red and blue traces above and below the barcode represent relative enrichment. P values for induced and repressed gene sets (indicating significant positive correlation with genes upregulated or downregulated upon Ikaros restoration respectively) are calculated separately using a mean-rank gene set test (2). Figure S5. (A) Spleens isolated from representative leukemic recipient mice showing regression of ALL65 and ALL211 following 7 days of Dox treatment, but progression of ALL11. Note that the spleen from the ALL11 day 3 Dox treated mouse is unusually small and likely reflects a lower leukemia burden at the start of Dox treatment for this particular mouse. (B-D) Kaplan- Meier survival curves for leukemic recipient mice bearing ALL65 (B), ALL211 (C), and ALL11 (D). Following disease establishment mice were either left untreated (black) or Dox treated. Negative (blue) indicates recipients of T-ALL cells that were either uninfected or
16 transduced with a negative control Cherry-expressing retroviral vector. Dox treatment significantly extended survival of mice bearing ALL65 (median survival for untreated and Doxtreated mice: 4 days vs 15 days respectively, P =.1, log rank test) and ALL211 (1 days vs 25 days, P =.1) but not ALL11 (7 days for both, P =.17). Recipients of T-ALL cells previously transduced with shikaros (red) or ICN1 (green) vectors promoted relapse of ALL65 and ALL211 as described in the text (transduced [shikaros or ICN1] vs negative: P <.5 and P <.1 for ALL65 and ALL211 respectively). (E) Western blot analysis of Ikaros expression in ALL65 and ALL11 cells isolated from representative leukemic mice that were untreated or Dox treated for 3 days. The two panels are cropped from the same blot, indicating similar Ikaros restoration in each leukemia. Figure S6. (A) Western blot analysis of Ikaros isoform expression in ALL65 leukemia cells isolated from independent leukemic mice that were either untreated (d), treated with Dox for 3 days (d3), or upon relapse following Dox treatment. #1 and #2 are leukemias isolated from two different relapsed mice after 29 days Dox. Asterisks indicate Ikaros species in relapsed leukemias not detected in untreated leukemias. Actin is a loading control. (B) Average Ikaros and Hes1 mrna expression detected by RT-qPCR in ALL65, ALL11, and ALL211, comparing mice that were untreated (d), treated with Dox for 3 days (d3), or upon relapse following Dox treatment. Datapoints represent leukemias isolated from independent mice. Hes1 expression in ALL65 is not significantly different in d vs relapse (P =.15, unpaired t test with Welch s correction). Figure S7. (A-C) RNA-seq track comparison of ALL65, ALL11, and ALL211 showing reads mapped to the Notch1 coding region altered by mutation in ALL65 (A), ALL11 (B), and ALL211 (C). Read numbers for each region are indicated on the right. (D) Sanger sequence electropherogram of a PCR product amplified from genomic DNA isolated from ALL211, showing a heterozygous S2446fsX1 frameshift mutation that introduces a premature stop codon in the Notch1 PEST domain. The same region amplified from ALL11 is shown as a wild type control. (E) Representative Sanger sequence electropherogram of a PCR product amplified from ALL11 cdna showing Notch1 mutations in trans with amino acid substititions highlighted in red. Figure S8. (A,B) Gene set analysis barcode plots of differential gene expression upon Ikaros restoration in T-ALL from a combined analysis of ALL65, ALL11, and ALL211. The barcode format is as described for Figure 2E. The Myc-regulated gene set in (A) comprises direct Mycinduced genes (red bars) or direct Myc-repressed genes (blue bars) identified previously in a
17 human B cell line harboring a tet-regulated Myc construct (3). The Myc cluster gene set in (B) comprises 84 genes with mrna expression closely correlated to Myc recently identified in detailed gene expression profiling of multiple stages of thymic T cell development (4). (C) Ikaros binding at the Myc locus in DP thymocytes. Grey bars below the Bio-ChIP-seq track indicate significant Ikaros-binding (P < 1 1 ). The Y axis indicates the number of mapped sequence reads. (D) Ikaros Bio-ChIP-seq track spanning the Myc locus and the Notch-driven Myc enhancer (N-Me) regulatory element (5, 6) ~1.3 Mb 3 of the Myc gene (arrowed), showing no binding of Ikaros to the N-Me in DP thymocytes. Figure S9. Western blot analysis of IKZF1 expression in the human T-ALL cell lines CUTLL1 and HPB-ALL following treatment with the GSI Compound E (CompE, 5 nm) for 24 and 48 h as indicated. ACTB (beta-actin) is a loading control.
18 Table S1. Gene expression changes upon acute Ikaros restoration in ALL11 - Top 1 genes RNA-Seq expression analysis of Vav-tTA;TRE-GFP-shIkaros leukemia #11 isolated from transplant recipient mice. 3 days doxycycline (GFPmid) vs. untreated (GFPhigh) INCREASED EXPRESSION Rank Symbol logfc AveLogCPM t PValue FDR 1 Ikzf E E-7 2 Cd E E-5 3 P2rx E E-5 4 Trac E E-5 5 Dusp E E-5 6 Limd E E-5 7 Cmtm E E-5 8 Shisa E E-5 9 Gsn E E-5 1 Dgka E E-5 11 Prr E E-5 12 Laptm E E-5 13 Kcnf E E-5 14 Igsf E E-5 15 Gpr E E-5 16 Jakmip E E-5 17 Stk E-7 8.9E-5 18 Tigit E-7 9.1E-5 19 Bzrap E-7 9.1E-5 2 Wdr E-7 9.1E-5 21 Egr E-7 9.1E-5 22 Lrp E-7 9.1E-5 23 Pld E-7 9.1E-5 24 Rab E-7 9.1E-5 25 Arid5a E-7 9.1E-5 26 Akna E-7 9.1E-5 27 Edem E-7 9.1E-5 28 Cd E-7 9.1E-5 29 Fam15a E-7 9.1E-5 3 Sp E-7 9.1E-5 31 Zfand E-7 9.1E-5 32 Cdh E-7 9.1E-5 33 Prkcq E-7 9.1E-5 34 Gem E-7 9.1E-5 35 Stk11ip E-7 9.1E-5 36 Itgam E-7 9.1E-5 37 Plcxd E-7 9.1E-5 38 Foxo E E-5 39 Gramd1a E-7 9.6E-5 4 Trav13n E E-5 41 Tor2a E E-5 42 Pstpip E-7 1.E-4 43 Pdlim E-7 1.E-4 44 Nlrc E-7 1.6E-4 45 Dennd1c E-7 1.6E-4 46 Gadd45a E-7 1.6E-4 47 Arhgap E-7 1.6E N6Rik E-7 1.6E-4 49 Aff E E-4 5 Rbl E E-4 51 Trav E E-4 52 Arid5b E-6 1.3E-4 53 Abhd E E-4 54 Nedd E E-4 55 Srp E E-4 56 Chmp1b E E-4 57 Zfp E E-4 58 Nbeal E E-4 59 Il12rb E E-4 6 Rasal E E-4 61 Arhgap E E-4 62 Rab E E-4 63 Flna E E-4 64 Pycard E E-4 65 Ifngr E E-4 66 Extl E E-4 67 Jak E E-4 68 Cdc42se E E-4 69 Ehd E E-4
19 7 Plgrkt E E-4 71 Hmha E E-4 72 Coro1a E-6 1.6E-4 73 Evi2b E-6 1.6E-4 74 Cd E-6 1.6E-4 75 Cd E-6 1.6E-4 76 Zbp E-6 1.6E-4 77 Rec E-6 1.6E-4 78 Fgd E-6 1.6E-4 79 Trpv E E-4 8 Cdyl E E-4 81 Fmnl E E-4 82 Nr4a E E-4 83 Trav13n E E-4 84 Trbv E E-4 85 Cd8a E E-4 86 Lcp E E-4 87 Xylt E E-4 88 Cd E E-4 89 Traj E E-4 9 P2ry E E-4 91 Celf E E-4 92 Gimap E-6 1.7E-4 93 Mtmr E E-4 94 Plekhg E E-4 95 Slc36a E E-4 96 Ephx E E-4 97 A4378G23Rik E E-4 98 Txnip E E-4 99 Adamts E E-4 1 Aes E E-4 DECREASED EXPRESSION Probe rank Symbol logfc AveExpr t P.Value FDR 1 Il9r E E-5 2 Tns E E-5 3 Maged E E-5 4 Mpzl E E-5 5 Slfn E E-5 6 Tbc1d E E-5 7 Dock E E-5 8 Elfn E E-5 9 Megf E E-5 1 Maoa E E-5 11 Tmed E-7 9.1E-5 12 C1qtnf E-7 9.1E-5 13 Polr1b E-7 9.1E-5 14 Tmem55a E-7 9.1E-5 15 Klf E-7 9.1E-5 16 Kifap E-7 9.1E-5 17 Pdxk E-7 9.1E-5 18 Eif4ebp E-7 9.1E-5 19 Wls E-7 9.1E-5 2 Matn E-7 9.1E-5 21 Slc45a E-7 9.1E-5 22 Qsox E-7 9.1E-5 23 BC E-7 9.1E-5 24 Spats E E-5 25 Wscd E-7 1.6E-4 26 Igfbpl E-7 1.6E-4 27 Myo5b E-7 1.7E-4 28 Myzap E E-4 29 Qser E E-4 3 Pkn E E G12Rik E E-4 32 Tspan E E-4 33 Igf1r E E-4 34 Hes E E-4 35 Irs E-6 1.3E-4 36 Uaca E-6 1.3E-4 37 C1qtnf E-6 1.3E-4 38 Vgll E-6 1.3E-4 39 Rsu E E-4 4 Rps6kl E E-4 41 Cluh E E-4
20 A12Rik E E-4 43 Tmem176b E E-4 44 Rab E E-4 45 Myh E E-4 46 Emp E E-4 47 Cdv E E-4 48 Arhgap E E-4 49 Gid E E-4 5 Carkd E E-4 51 Kdelc E E-4 52 Nadk E E-4 53 Klhl E E-4 54 Pdgfrb E E-4 55 Psrc E-6 1.6E-4 56 Vash E-6 1.6E-4 57 Mtf E-6 1.6E-4 58 Mrps E E-4 59 Itgb E E-4 6 Hivep E E-4 61 Epb4.1l E E-4 62 Mgat4b E E-4 63 Acvr E E-4 64 Ahctf E E-4 65 Frmd4a E E-4 66 Npr E E-4 67 D8331N3Rik E E-4 68 Tmem132a E E-4 69 Utp E E-4 7 Arhgap E E-4 71 Ctnnd E E-4 72 Ccdc E E-4 73 Nat E E-4 74 Bst E E-4 75 Rab E E-4 76 Fam188b E E-4 77 Josd E E-4 78 Aldh1b E E-4 79 Tfap E E-4 8 Anks E E-4 81 Zfp E E-4 82 Mettl E E-4 83 Nt5dc E E-4 84 Trip E E-4 85 Dpy19l E-6 2.1E-4 86 Jazf E-6 2.1E-4 87 Akap E-6 2.1E-4 88 Rnpep E-6 2.2E-4 89 Gphn E-6 2.9E-4 9 Wdr E-6 2.9E-4 91 Podnl E E-4 92 Tnfrsf E E-4 93 Vcl E E-4 94 Ccdc E E-4 95 Bmp E E-4 96 Bsn E E-4 97 Vwa E E-4 98 Cpne E E-4 99 Tnfsf E E-4 1 Smo E E-4
21 Table S2. Ikaros activated genes Activated upon 3d Dox treatment in ALL65, ALL11and ALL211 - sorted by FDR Rank GeneID Symbol Name ChIP logfc AveExpr t P.Value FDR Gimap3 GTPase, IMAP family member 3 TRUE E E Gramd3 GRAM domain containing 3 TRUE E E Ecm1 extracellular matrix protein 1 FALSE E E Gimap6 GTPase, IMAP family member 6 TRUE E E N6Rik RIKEN cdna N6 gene FALSE E E Itgb7 integrin beta 7 FALSE E E Ikzf1 IKAROS family zinc finger 1 TRUE E E Phf11b PHD finger protein 11B FALSE E E Ehd3 EH-domain containing 3 TRUE E E Cd7 CD7 antigen FALSE E E Gimap4 GTPase, IMAP family member 4 FALSE E E Pik3r3 phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) TRUE E E B6319A1Rik RIKEN cdna B6319A1 gene FALSE E E Rab37 RAB37, member of RAS oncogene family TRUE E E Vcam1 vascular cell adhesion molecule 1 FALSE E E Shisa5 shisa homolog 5 (Xenopus laevis) FALSE E E Gimap7 GTPase, IMAP family member 7 FALSE E E S1pr1 sphingosine-1-phosphate receptor 1 FALSE E E Cd37 CD37 antigen FALSE E E Klf2 Kruppel-like factor 2 (lung) TRUE E E Ephx1 epoxide hydrolase 1, microsomal FALSE E E Itgb3 integrin beta 3 FALSE E E Acoxl acyl-coenzyme A oxidase-like TRUE E E Slfn5 schlafen 5 TRUE E E Ifi27l2a interferon, alpha-inducible protein 27 like 2A FALSE E-5 1.9E Wdr1 WD repeat domain 1 FALSE E E Ubash3b ubiquitin associated and SH3 domain containing, B TRUE E E Cmtm6 CKLF-like MARVEL transmembrane domain containing 6 FALSE E E Gimap8 GTPase, IMAP family member 8 TRUE E E Cd53 CD53 antigen FALSE E E Trp53inp2 transformation related protein 53 inducible nuclear protein 2 FALSE E E Ampd1 adenosine monophosphate deaminase 1 FALSE E E Pde6a phosphodiesterase 6A, cgmp-specific, rod, alpha TRUE E E Flt3l FMS-like tyrosine kinase 3 ligand TRUE E E Dgka diacylglycerol kinase, alpha TRUE E E Cd44 CD44 antigen TRUE E-4 2.7E Hsd11b1 hydroxysteroid 11-beta dehydrogenase 1 TRUE E-4 2.7E Fas Fas (TNF receptor superfamily member 6) TRUE E-4 2.7E Vim vimentin TRUE E-4 2.9E Flna filamin, alpha FALSE E-4 2.9E Pacsin1 protein kinase C and casein kinase substrate in neurons 1 FALSE E-4 2.9E Fcna ficolin A FALSE E-4 2.9E Cdc14b CDC14 cell division cycle 14B FALSE E-4 2.9E Fcgrt Fc receptor, IgG, alpha chain transporter FALSE E-4 2.9E Gm11738 predicted gene FALSE E-4 2.9E Gm8995 predicted gene 8995 FALSE E E Slc4a1 solute carrier family 4 (anion exchanger), member 1 FALSE E E Spats2l spermatogenesis associated, serine-rich 2-like TRUE E E Foxo1 forkhead box O1 TRUE E E Slc43a2 solute carrier family 43, member 2 TRUE E E Sema4a sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A FALSE E E Itgb2 integrin beta 2 TRUE E E Zbp1 Z-DNA binding protein 1 FALSE E E Aff4 AF4/FMR2 family, member 4 TRUE E E P2rx7 purinergic receptor P2X, ligand-gated ion channel, 7 TRUE E E Gm2559 predicted gene, 2559 FALSE E E Celf2 CUGBP, Elav-like family member 2 TRUE E E Cpeb1 cytoplasmic polyadenylation element binding protein 1 FALSE E E Nsg2 neuron specific gene family member 2 TRUE E E Glcci1 glucocorticoid induced transcript 1 TRUE E E Tlr1 toll-like receptor 1 FALSE E E Pdlim4 PDZ and LIM domain 4 FALSE E E Pyhin1 pyrin and HIN domain family, member 1 FALSE E E A4378G23Rik RIKEN cdna A4378G23 gene TRUE E-4 2.2E Arid5b AT rich interactive domain 5B (MRF1-like) TRUE E-4 2.2E Cytip cytohesin 1 interacting protein TRUE E-4 2.2E Gem GTP binding protein (gene overexpressed in skeletal muscle) FALSE E-4 2.2E Nfkbiz nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta TRUE E-4 2.2E Ccr7 chemokine (C-C motif) receptor 7 TRUE E-4 2.2E Itgam integrin alpha M TRUE E-4 2.2E Rrad Ras-related associated with diabetes FALSE E-4 2.2E Ankrd44 ankyrin repeat domain 44 TRUE E-4 2.2E Slc28a2 solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 FALSE E-4 2.2E Kdm7a lysine (K)-specific demethylase 7A FALSE E-4 2.2E Cysltr2 cysteinyl leukotriene receptor 2 TRUE E-4 2.2E Hc hemolytic complement FALSE E-4 2.2E Tor2a torsin family 2, member A FALSE E-4 2.2E Gsn gelsolin FALSE E-4 2.2E Pde4d phosphodiesterase 4D, camp specific TRUE E-4 2.2E Pced1b PC-esterase domain containing 1B TRUE E-4 2.2E Nedd9 neural precursor cell expressed, developmentally down-regulated gene 9 TRUE E-4 2.2E Prkcq protein kinase C, theta TRUE E-4 2.2E Cdc42se1 CDC42 small effector 1 TRUE E-4 2.2E Ap3m2 adaptor-related protein complex 3, mu 2 subunit TRUE E-4 2.2E Glipr1 GLI pathogenesis-related 1 (glioma) FALSE E-4 2.2E Sidt1 SID1 transmembrane family, member 1 FALSE E-4 2.2E Dnah17 dynein, axonemal, heavy chain 17 FALSE E-4 2.2E Fam134b family with sequence similarity 134, member B FALSE E-4 2.2E Limd2 LIM domain containing 2 TRUE E-4 2.2E Il21r interleukin 21 receptor TRUE E E Sp1 nuclear antigen Sp1 FALSE E E L24Rik RIKEN cdna L24 gene TRUE E E Arl4c ADP-ribosylation factor-like 4C TRUE E-4 2.3E Themis thymocyte selection associated TRUE E-4 2.3E Tmsb1 thymosin, beta 1 TRUE E E Hbb-bs hemoglobin, beta adult s chain FALSE E E Phf11c PHD finger protein 11C FALSE E E Bcl3 B cell leukemia/lymphoma 3 FALSE E E Tnfaip8l2 tumor necrosis factor, alpha-induced protein 8-like 2 TRUE E E Wnt8b wingless related MMTV integration site 8b FALSE E E Samd9l sterile alpha motif domain containing 9-like FALSE E E Rbms1 RNA binding motif, single stranded interacting protein 1 FALSE E E Add3 adducin 3 (gamma) FALSE E E Zfp36 zinc finger protein 36 TRUE E E Ikzf2 IKAROS family zinc finger 2 TRUE E E Laptm5 lysosomal-associated protein transmembrane 5 TRUE E E Trav13n-4 T cell receptor alpha variable 13N-4 FALSE E E Ddit4 DNA-damage-inducible transcript 4 FALSE E E Cxcr6 chemokine (C-X-C motif) receptor 6 FALSE E E Cd52 CD52 antigen TRUE E E-2
22 Cd6 CD6 antigen TRUE E E Dennd3 DENN/MADD domain containing 3 FALSE E E Pld3 phospholipase D family, member 3 TRUE E E Gimap5 GTPase, IMAP family member 5 TRUE E E Ikbke inhibitor of kappab kinase epsilon TRUE E E Btg2 B cell translocation gene 2, anti-proliferative TRUE E E BC94916 cdna sequence BC94916 FALSE E E St3gal2 ST3 beta-galactoside alpha-2,3-sialyltransferase 2 TRUE E E Lpar6 lysophosphatidic acid receptor 6 TRUE E E Pglyrp2 peptidoglycan recognition protein 2 TRUE E-4 2.5E Prr13 proline rich 13 TRUE E-4 2.5E Fam214a family with sequence similarity 214, member A FALSE E E Il6ra interleukin 6 receptor, alpha TRUE E E Evi2b ecotropic viral integration site 2b FALSE E E Tgfb3 transforming growth factor, beta 3 FALSE E E Pkp3 plakophilin 3 FALSE E E Rhof ras homolog gene family, member f TRUE E E Il4ra interleukin 4 receptor, alpha TRUE E E Adam12 a disintegrin and metallopeptidase domain 12 (meltrin alpha) FALSE E E Gpr65 G-protein coupled receptor 65 FALSE E E H2-Ob histocompatibility 2, O region beta locus FALSE E E C1qc complement component 1, q subcomponent, C chain FALSE E E Cotl1 coactosin-like 1 (Dictyostelium) TRUE E E St6gal1 beta galactoside alpha 2,6 sialyltransferase 1 TRUE E E Cd226 CD226 antigen TRUE E E Slc46a3 solute carrier family 46, member 3 FALSE E E Batf basic leucine zipper transcription factor, ATF-like TRUE E-4 2.7E Wdr78 WD repeat domain 78 TRUE E-4 2.7E Rasgrp1 RAS guanyl releasing protein 1 FALSE E-4 2.7E Wdr44 WD repeat domain 44 TRUE E-4 2.7E Slamf1 signaling lymphocytic activation molecule family member 1 TRUE E-4 2.7E Cd5l CD5 antigen-like FALSE E-4 2.7E Tnik TRAF2 and NCK interacting kinase TRUE E-4 2.7E Cmya5 cardiomyopathy associated 5 TRUE E E Tuba1a tubulin, alpha 1A TRUE E E Sepw1 selenoprotein W, muscle 1 FALSE E E Il12rb1 interleukin 12 receptor, beta 1 TRUE E E Gadd45a growth arrest and DNA-damage-inducible 45 alpha TRUE E E Gimap1 GTPase, IMAP family member 1 TRUE E E Ccng2 cyclin G2 TRUE E E Chmp1b charged multivesicular body protein 1B FALSE E E Atp1b1 ATPase, Na+/K+ transporting, beta 1 polypeptide TRUE E E Trim12a tripartite motif-containing 12A FALSE E E Gm11346 predicted gene TRUE E E Gxylt1 glucoside xylosyltransferase 1 FALSE E E Ndrg3 N-myc downstream regulated gene 3 TRUE E E Rgs14 regulator of G-protein signaling 14 FALSE E E Hmox1 heme oxygenase (decycling) 1 TRUE E E Cdkn2d cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) FALSE E E Emp3 epithelial membrane protein 3 TRUE E E Fam15a family with sequence similarity 15, member A TRUE E E Slfn1 schlafen 1 FALSE E E Pik3r1 phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) TRUE E E Ypel3 yippee-like 3 (Drosophila) FALSE E E Ltb lymphotoxin B FALSE E E Mxd4 Max dimerization protein 4 FALSE E E Nod1 nucleotide-binding oligomerization domain containing 1 TRUE E E Tmc8 transmembrane channel-like gene family 8 TRUE E E Inpp4a inositol polyphosphate-4-phosphatase, type I FALSE E E Arhgap25 Rho GTPase activating protein 25 TRUE E E Treml2 triggering receptor expressed on myeloid cells-like 2 TRUE E E Dap death-associated protein TRUE E E Dyrk1b dual-specificity tyrosine-(y)-phosphorylation regulated kinase 1b FALSE E E Lysmd3 LysM, putative peptidoglycan-binding, domain containing 3 FALSE E E Esyt1 extended synaptotagmin-like protein 1 TRUE E E Sirpa signal-regulatory protein alpha FALSE E E Slc37a3 solute carrier family 37 (glycerol-3-phosphate transporter), member 3 TRUE E E A6333H2Rik RIKEN cdna A6333H2 gene FALSE E E Cnr2 cannabinoid receptor 2 (macrophage) TRUE E E Nbeal2 neurobeachin-like 2 TRUE E E Gm14446 predicted gene FALSE E E Jup junction plakoglobin FALSE E E Ptpn22 protein tyrosine phosphatase, non-receptor type 22 (lymphoid) TRUE E E Fam12a family with sequence similarity 12, member A TRUE E E Srp14 signal recognition particle 14 TRUE E E Dmbt1 deleted in malignant brain tumors 1 FALSE E E Pstpip1 proline-serine-threonine phosphatase-interacting protein 1 TRUE E E A932I21Rik RIKEN cdna A932I21 gene FALSE E E Pdlim2 PDZ and LIM domain 2 FALSE E E Gramd1a GRAM domain containing 1A TRUE E E Sesn3 sestrin 3 FALSE E E Stk17b serine/threonine kinase 17b (apoptosis-inducing) TRUE E E Lcp2 lymphocyte cytosolic protein 2 TRUE E E Trp53inp1 transformation related protein 53 inducible nuclear protein 1 TRUE E E Rinl Ras and Rab interactor-like FALSE E E Ms4a4b membrane-spanning 4-domains, subfamily A, member 4B FALSE E E Lipa lysosomal acid lipase A TRUE E E Gm14718 predicted gene FALSE E E Grap GRB2-related adaptor protein TRUE E E Skap1 src family associated phosphoprotein 1 FALSE E E Man1a mannosidase 1, alpha TRUE E E Prf1 perforin 1 (pore forming protein) FALSE E E Coro1a coronin, actin binding protein 1A TRUE E E Slc26a2 solute carrier family 26 (sulfate transporter), member 2 FALSE E E Ccdc64 coiled-coil domain containing 64 TRUE E E Lysmd2 LysM, putative peptidoglycan-binding, domain containing 2 FALSE E E Plgrkt plasminogen receptor, C-terminal lysine transmembrane protein FALSE E E Pgap1 post-gpi attachment to proteins 1 TRUE E E Sdc3 syndecan 3 FALSE E E Nr4a1 nuclear receptor subfamily 4, group A, member 1 TRUE E E Arhgef18 rho/rac guanine nucleotide exchange factor (GEF) 18 FALSE E E Dleu2 deleted in lymphocytic leukemia, 2 TRUE E E Rap1b RAS related protein 1b TRUE E E Suox sulfite oxidase FALSE E E Capn3 calpain 3 TRUE E E Il16 interleukin 16 TRUE E E Tspan13 tetraspanin 13 TRUE E E Runx2 runt related transcription factor 2 TRUE E E Ypel5 yippee-like 5 (Drosophila) TRUE E E Ankrd5 ankyrin repeat domain 5 TRUE E E Cpeb4 cytoplasmic polyadenylation element binding protein 4 TRUE E E Sla2 Src-like-adaptor 2 TRUE E E Atxn1 ataxin 1 TRUE E E Tnfrsf14 tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) TRUE E E-2
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks. Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks
 Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Single-site association results for 136 SCARB1 genotyped variants with HDL-C.
 Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
High mobility group 1 HMG1
 Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
TABLE OF CONTENTS Page
 TABLE OF CONTENTS Page Acknowledgements... VI Declaration... VII List of abbreviations... VIII 1. INTRODUCTION... 1 2. BASIC PRINCIPLES... 3 2.1. Hereditary Colorectal Cancer... 3 2.1.1. Differential diagnosis...
TABLE OF CONTENTS Page Acknowledgements... VI Declaration... VII List of abbreviations... VIII 1. INTRODUCTION... 1 2. BASIC PRINCIPLES... 3 2.1. Hereditary Colorectal Cancer... 3 2.1.1. Differential diagnosis...
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
HOMEWORK#1. t E(x) = 1 λ = (b) Find the median lifetime of a randomly selected light bulb. Answer:
 HOMEWORK# 52258 李亞晟 Eercise 2. The lifetime of light bulbs follows an eponential distribution with a hazard rate of. failures per hour of use (a) Find the mean lifetime of a randomly selected light bulb.
HOMEWORK# 52258 李亞晟 Eercise 2. The lifetime of light bulbs follows an eponential distribution with a hazard rate of. failures per hour of use (a) Find the mean lifetime of a randomly selected light bulb.
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
Biostatistics for Health Sciences Review Sheet
 Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplementary Information Titles
 Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ. Σπίγγος Γεώργιος
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Abstract... I. Zusammenfassung... II. 1 Aim of the work Introduction Short overview of Chinese hamster ovary cell lines...
 III Contents I II III Abstract... I Zusammenfassung... II Contents... III 1 Aim of the work... 1 2 Introduction... 2 2.1 Short overview of Chinese hamster ovary cell lines... 2 2.2 Unspecific mutations:
III Contents I II III Abstract... I Zusammenfassung... II Contents... III 1 Aim of the work... 1 2 Introduction... 2 2.1 Short overview of Chinese hamster ovary cell lines... 2 2.2 Unspecific mutations:
Supporting Information. Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka
 Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3
 Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Chapter 1. Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes
 Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Mouse Gene 2.0 ST Array Shn3. Runx2 Shn3. Runx2 Shn3 Runx2. adult. (Schnurri, Shn)3/Hivep3. Runx2 Shn/Hivep. ST2 BMP-2 Shn3
 adult (Schnurri, Shn)3/Hivep3 Runx2 Shn/Hivep Shn3 Shn3 Shn3 Shn3 Shn1 Shn2 Shn3 gain-of-function loss-of-function Shn3 in vivo mimic MC3T3-E1 ST-2 ATDC5 C3H10T1/2 ALP RT-PCR alcian blue RT-PCR Affymetrix
adult (Schnurri, Shn)3/Hivep3 Runx2 Shn/Hivep Shn3 Shn3 Shn3 Shn3 Shn1 Shn2 Shn3 gain-of-function loss-of-function Shn3 in vivo mimic MC3T3-E1 ST-2 ATDC5 C3H10T1/2 ALP RT-PCR alcian blue RT-PCR Affymetrix
Matrices and vectors. Matrix and vector. a 11 a 12 a 1n a 21 a 22 a 2n A = b 1 b 2. b m. R m n, b = = ( a ij. a m1 a m2 a mn. def
 Matrices and vectors Matrix and vector a 11 a 12 a 1n a 21 a 22 a 2n A = a m1 a m2 a mn def = ( a ij ) R m n, b = b 1 b 2 b m Rm Matrix and vectors in linear equations: example E 1 : x 1 + x 2 + 3x 4 =
Matrices and vectors Matrix and vector a 11 a 12 a 1n a 21 a 22 a 2n A = a m1 a m2 a mn def = ( a ij ) R m n, b = b 1 b 2 b m Rm Matrix and vectors in linear equations: example E 1 : x 1 + x 2 + 3x 4 =
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver
 Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Guo_Fig. S1. Atg7 +/+ Atg7 -/- FBP_M+6 DHAP_M+3. Glucose Uptake Rate G6P_M+6. nmol/ul cell/hr
 Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Daewoo Technopark A-403, Dodang-dong, Wonmi-gu, Bucheon-city, Gyeonggido, Korea LM-80 Test Report
 LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
Instruction Execution Times
 1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
ΠΣΤΧΛΑΚΘ ΕΡΓΑΛΑ ΜΕΣΡΘΕΛ ΟΠΣΛΚΟΤ ΒΑΚΟΤ ΑΣΜΟΦΑΛΡΑ ΜΕ ΘΛΛΑΚΟ ΦΩΣΟΜΕΣΡΟ ΕΚΟ
 ln(f( )) ΑΡΛΣΟΣΕΛΕΛΟ ΠΑΝΕΠΛΣΘΜΛΟ ΚΕΑΛΟΝΛΚΘ ΣΜΘΜΑ ΦΤΛΚΘ ΠΣΤΧΛΑΚΘ ΕΡΓΑΛΑ ΜΕΣΡΘΕΛ ΟΠΣΛΚΟΤ ΒΑΚΟΤ ΑΣΜΟΦΑΛΡΑ ΜΕ ΘΛΛΑΚΟ ΦΩΣΟΜΕΣΡΟ ΕΚΟ 4,6 4,4 4,2 4,0 3,8 3,6 3,4 3,2 3,0 2,8 2,6 2,4 2,2 2,0 1,8 1,6 1 2 3 4 5
ln(f( )) ΑΡΛΣΟΣΕΛΕΛΟ ΠΑΝΕΠΛΣΘΜΛΟ ΚΕΑΛΟΝΛΚΘ ΣΜΘΜΑ ΦΤΛΚΘ ΠΣΤΧΛΑΚΘ ΕΡΓΑΛΑ ΜΕΣΡΘΕΛ ΟΠΣΛΚΟΤ ΒΑΚΟΤ ΑΣΜΟΦΑΛΡΑ ΜΕ ΘΛΛΑΚΟ ΦΩΣΟΜΕΣΡΟ ΕΚΟ 4,6 4,4 4,2 4,0 3,8 3,6 3,4 3,2 3,0 2,8 2,6 2,4 2,2 2,0 1,8 1,6 1 2 3 4 5
Aluminum Electrolytic Capacitors
 Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors (Large Can Type)
 Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Aronia. melanocarpa. Επιβλεπονηερ καθηγηηερ:
 Αλεξάνδπειο Τεσνολογικό Εκπαιδεςηικό Ίδπςμα Θεζζαλονίκηρ Σσολή Τεσνολογίαρ Γεωπονίαρ Και Τεσνολογίαρ Τποθίμων Διαηποθήρ Aronia melanocarpa Αξιολόγηζη ηηρ επίδπαζηρ Ελληνικών απομονώζεων ηος γένοςρ Trichoderma
Αλεξάνδπειο Τεσνολογικό Εκπαιδεςηικό Ίδπςμα Θεζζαλονίκηρ Σσολή Τεσνολογίαρ Γεωπονίαρ Και Τεσνολογίαρ Τποθίμων Διαηποθήρ Aronia melanocarpa Αξιολόγηζη ηηρ επίδπαζηρ Ελληνικών απομονώζεων ηος γένοςρ Trichoderma
EE512: Error Control Coding
 EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ Παθοφυσιολογία ΙΙ Ογκολογία Υπεύθυνος μαθήματος: Καθηγητής Αλέξανδρος Α. Δρόσος Άδειες Χρήσης Το παρόν εκπαιδευτικό υλικό υπόκειται σε άδειες χρήσης Creative
ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ Παθοφυσιολογία ΙΙ Ογκολογία Υπεύθυνος μαθήματος: Καθηγητής Αλέξανδρος Α. Δρόσος Άδειες Χρήσης Το παρόν εκπαιδευτικό υλικό υπόκειται σε άδειες χρήσης Creative
ΜΟΡΙΑΚΕΣ ΤΕΧΝΙΚΕΣ ΠΡΑΓΜΑΤΙΚΟΥ ΧΡΟΝΟΥ ΣΠΑΝΑΚΗΣ ΝΙΚΟΣ
 ΜΟΡΙΑΚΕΣ ΤΕΧΝΙΚΕΣ ΠΡΑΓΜΑΤΙΚΟΥ ΧΡΟΝΟΥ ΣΠΑΝΑΚΗΣ ΝΙΚΟΣ Αρχή λειτουργίας Real-time PCR * Βασίζεται στην ανίχνευση και ποσοτικοποίηση του φθορισμού που εκπέμπεται από ειδικά φθοριοχρώματα * Η αρχική αύξηση
ΜΟΡΙΑΚΕΣ ΤΕΧΝΙΚΕΣ ΠΡΑΓΜΑΤΙΚΟΥ ΧΡΟΝΟΥ ΣΠΑΝΑΚΗΣ ΝΙΚΟΣ Αρχή λειτουργίας Real-time PCR * Βασίζεται στην ανίχνευση και ποσοτικοποίηση του φθορισμού που εκπέμπεται από ειδικά φθοριοχρώματα * Η αρχική αύξηση
Bayesian statistics. DS GA 1002 Probability and Statistics for Data Science.
 Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ ΡΑΣΗ ΤΟΥ ΓΑΣΤΡΙΚΟΥ ΒΛΕΝΝΟΓΟΝΟΥ ΣΤΗ ΛΟΙΜΩΞΗ ΜΕ ΕΛΙΚΟΒΑΚΤΗΡΙ ΙΟ ΤΟΥ ΠΥΛΩΡΟΥ ΠΡΙΝ ΚΑΙ ΜΕΤΑ ΤΗ ΘΕΡΑΠΕΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
«Συντήρηση αχλαδιών σε νερό. υπό την παρουσία σπόρων σιναπιού (Sinapis arvensis).»
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine
 Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Code Breaker. TEACHER s NOTES
 TEACHER s NOTES Time: 50 minutes Learning Outcomes: To relate the genetic code to the assembly of proteins To summarize factors that lead to different types of mutations To distinguish among positive,
TEACHER s NOTES Time: 50 minutes Learning Outcomes: To relate the genetic code to the assembly of proteins To summarize factors that lead to different types of mutations To distinguish among positive,
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 12/06/2013 Αρ. πρωτ. 4972 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 12/06/2013 Αρ. πρωτ. 4972 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
ΖΩΝΟΠΟΙΗΣΗ ΤΗΣ ΚΑΤΟΛΙΣΘΗΤΙΚΗΣ ΕΠΙΚΙΝΔΥΝΟΤΗΤΑΣ ΣΤΟ ΟΡΟΣ ΠΗΛΙΟ ΜΕ ΤΗ ΣΥΜΒΟΛΗ ΔΕΔΟΜΕΝΩΝ ΣΥΜΒΟΛΟΜΕΤΡΙΑΣ ΜΟΝΙΜΩΝ ΣΚΕΔΑΣΤΩΝ
 EΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΕΙΟ Τμήμα Μηχανικών Μεταλλείων-Μεταλλουργών ΖΩΝΟΠΟΙΗΣΗ ΤΗΣ ΚΑΤΟΛΙΣΘΗΤΙΚΗΣ ΕΠΙΚΙΝΔΥΝΟΤΗΤΑΣ ΜΕ ΤΗ ΣΥΜΒΟΛΗ ΔΕΔΟΜΕΝΩΝ ΣΥΜΒΟΛΟΜΕΤΡΙΑΣ ΜΟΝΙΜΩΝ ΣΚΕΔΑΣΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Κιτσάκη Μαρίνα
EΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΕΙΟ Τμήμα Μηχανικών Μεταλλείων-Μεταλλουργών ΖΩΝΟΠΟΙΗΣΗ ΤΗΣ ΚΑΤΟΛΙΣΘΗΤΙΚΗΣ ΕΠΙΚΙΝΔΥΝΟΤΗΤΑΣ ΜΕ ΤΗ ΣΥΜΒΟΛΗ ΔΕΔΟΜΕΝΩΝ ΣΥΜΒΟΛΟΜΕΤΡΙΑΣ ΜΟΝΙΜΩΝ ΣΚΕΔΑΣΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Κιτσάκη Μαρίνα
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
Sampling Basics (1B) Young Won Lim 9/21/13
 Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων. Εξάμηνο 7 ο
 Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων Εξάμηνο 7 ο Procedures and Functions Stored procedures and functions are named blocks of code that enable you to group and organize a series of SQL and PL/SQL
Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων Εξάμηνο 7 ο Procedures and Functions Stored procedures and functions are named blocks of code that enable you to group and organize a series of SQL and PL/SQL
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΜΑΡΙΑΣ ΦΩΤΙΟΥ ΠΤΥΧΙΟΥΧΟΥ ΓΕΩΠΟΝΟΥ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΜΑΡΙΑΣ ΦΩΤΙΟΥ ΠΤΥΧΙΟΥΧΟΥ ΓΕΩΠΟΝΟΥ Συγκέντρωση των ελεύθερων αµινοξέων στο αµνιακό υγρό σε σχέση µε την εβδοµάδα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΜΑΡΙΑΣ ΦΩΤΙΟΥ ΠΤΥΧΙΟΥΧΟΥ ΓΕΩΠΟΝΟΥ Συγκέντρωση των ελεύθερων αµινοξέων στο αµνιακό υγρό σε σχέση µε την εβδοµάδα
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
2 Composition. Invertible Mappings
 Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
ANSWERSHEET (TOPIC = DIFFERENTIAL CALCULUS) COLLECTION #2. h 0 h h 0 h h 0 ( ) g k = g 0 + g 1 + g g 2009 =?
 Teko Classes IITJEE/AIEEE Maths by SUHAAG SIR, Bhopal, Ph (0755) 3 00 000 www.tekoclasses.com ANSWERSHEET (TOPIC DIFFERENTIAL CALCULUS) COLLECTION # Question Type A.Single Correct Type Q. (A) Sol least
Teko Classes IITJEE/AIEEE Maths by SUHAAG SIR, Bhopal, Ph (0755) 3 00 000 www.tekoclasses.com ANSWERSHEET (TOPIC DIFFERENTIAL CALCULUS) COLLECTION # Question Type A.Single Correct Type Q. (A) Sol least
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΑ ΤΜΗΜΑ ΝΑΥΤΙΛΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ ΝΑΥΤΙΛΙΑ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΑ ΤΜΗΜΑ ΝΑΥΤΙΛΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ ΝΑΥΤΙΛΙΑ ΝΟΜΙΚΟ ΚΑΙ ΘΕΣΜΙΚΟ ΦΟΡΟΛΟΓΙΚΟ ΠΛΑΙΣΙΟ ΚΤΗΣΗΣ ΚΑΙ ΕΚΜΕΤΑΛΛΕΥΣΗΣ ΠΛΟΙΟΥ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ που υποβλήθηκε στο
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΑ ΤΜΗΜΑ ΝΑΥΤΙΛΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ ΝΑΥΤΙΛΙΑ ΝΟΜΙΚΟ ΚΑΙ ΘΕΣΜΙΚΟ ΦΟΡΟΛΟΓΙΚΟ ΠΛΑΙΣΙΟ ΚΤΗΣΗΣ ΚΑΙ ΕΚΜΕΤΑΛΛΕΥΣΗΣ ΠΛΟΙΟΥ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ που υποβλήθηκε στο
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supporting Information
 Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
NMBTC.COM /
 Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ ΩΝ ΚΑΤΑ ΤΗΝ ΕΜΒΡΥΪΚΗ ΑΝΑΠΤΥΞΗ
 ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1
 Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Repeated measures Επαναληπτικές μετρήσεις
 ΠΡΟΒΛΗΜΑ Στο αρχείο δεδομένων diavitis.sav καταγράφεται η ποσότητα γλυκόζης στο αίμα 10 ασθενών στην αρχή της χορήγησης μιας θεραπείας, μετά από ένα μήνα και μετά από δύο μήνες. Μελετήστε την επίδραση
ΠΡΟΒΛΗΜΑ Στο αρχείο δεδομένων diavitis.sav καταγράφεται η ποσότητα γλυκόζης στο αίμα 10 ασθενών στην αρχή της χορήγησης μιας θεραπείας, μετά από ένα μήνα και μετά από δύο μήνες. Μελετήστε την επίδραση
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
519.22(07.07) 78 : ( ) /.. ; c (07.07) , , 2008
 .. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
.. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
ΕΚΤΙΜΗΣΗ ΤΟΥ ΚΟΣΤΟΥΣ ΤΩΝ ΟΔΙΚΩΝ ΑΤΥΧΗΜΑΤΩΝ ΚΑΙ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΑΡΑΓΟΝΤΩΝ ΕΠΙΡΡΟΗΣ ΤΟΥ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΕΚΤΙΜΗΣΗ ΤΟΥ ΚΟΣΤΟΥΣ ΤΩΝ ΟΔΙΚΩΝ ΑΤΥΧΗΜΑΤΩΝ ΚΑΙ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΑΡΑΓΟΝΤΩΝ ΕΠΙΡΡΟΗΣ ΤΟΥ ΔΙΑΤΡΙΒΗ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ ΔΗΜΗΤΡΙΟΥ Ν. ΠΙΤΕΡΟΥ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΕΚΤΙΜΗΣΗ ΤΟΥ ΚΟΣΤΟΥΣ ΤΩΝ ΟΔΙΚΩΝ ΑΤΥΧΗΜΑΤΩΝ ΚΑΙ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΑΡΑΓΟΝΤΩΝ ΕΠΙΡΡΟΗΣ ΤΟΥ ΔΙΑΤΡΙΒΗ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ ΔΗΜΗΤΡΙΟΥ Ν. ΠΙΤΕΡΟΥ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΤΕΧΝΟΛΟΓΙΑΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΥΠΟΛΟΓΙΣΤΩΝ Σύστημα Διαχείρισης Διαχρονικών Δεδομένων για Γονίδια ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΤΕΧΝΟΛΟΓΙΑΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΥΠΟΛΟΓΙΣΤΩΝ Σύστημα Διαχείρισης Διαχρονικών Δεδομένων για Γονίδια ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
Supplementary Materials for. Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides
 Supplementary Materials for Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides Indrek Kalvet, a Karl J. Bonney, a and Franziska Schoenebeck a * a Institute of Organic
Supplementary Materials for Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides Indrek Kalvet, a Karl J. Bonney, a and Franziska Schoenebeck a * a Institute of Organic
ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ
 ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Τομέας Περιβαλλοντικής Υδραυλικής και Γεωπεριβαλλοντικής Μηχανικής (III) Εργαστήριο Γεωπεριβαλλοντικής Μηχανικής TECHNICAL UNIVERSITY OF CRETE SCHOOL of
ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Τομέας Περιβαλλοντικής Υδραυλικής και Γεωπεριβαλλοντικής Μηχανικής (III) Εργαστήριο Γεωπεριβαλλοντικής Μηχανικής TECHNICAL UNIVERSITY OF CRETE SCHOOL of
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Statistical Inference I Locally most powerful tests
 Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
Numerical Analysis FMN011
 Numerical Analysis FMN011 Carmen Arévalo Lund University carmen@maths.lth.se Lecture 12 Periodic data A function g has period P if g(x + P ) = g(x) Model: Trigonometric polynomial of order M T M (x) =
Numerical Analysis FMN011 Carmen Arévalo Lund University carmen@maths.lth.se Lecture 12 Periodic data A function g has period P if g(x + P ) = g(x) Model: Trigonometric polynomial of order M T M (x) =
Μειέηε, θαηαζθεπή θαη πξνζνκνίσζε ηεο ιεηηνπξγίαο κηθξήο αλεκνγελλήηξηαο αμνληθήο ξνήο ΓΗΠΛΩΜΑΣΗΚΖ ΔΡΓΑΗΑ
 Μειέηε, θαηαζθεπή θαη πξνζνκνίσζε ηεο ιεηηνπξγίαο κηθξήο αλεκνγελλήηξηαο αμνληθήο ξνήο ΓΗΠΛΩΜΑΣΗΚΖ ΔΡΓΑΗΑ Κνηζακπφπνπινο Υ. Παλαγηψηεο Δπηβιέπσλ: Νηθφιανο Υαηδεαξγπξίνπ Καζεγεηήο Δ.Μ.Π Αζήλα, Μάξηηνο 2010
Μειέηε, θαηαζθεπή θαη πξνζνκνίσζε ηεο ιεηηνπξγίαο κηθξήο αλεκνγελλήηξηαο αμνληθήο ξνήο ΓΗΠΛΩΜΑΣΗΚΖ ΔΡΓΑΗΑ Κνηζακπφπνπινο Υ. Παλαγηψηεο Δπηβιέπσλ: Νηθφιανο Υαηδεαξγπξίνπ Καζεγεηήο Δ.Μ.Π Αζήλα, Μάξηηνο 2010
FORMULAS FOR STATISTICS 1
 FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
Nature Immunology: doi: /ni Supplementary Figure 1. Foxm1 loss leads to bone marrow hypocellularity.
 Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής
 Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Congruence Classes of Invertible Matrices of Order 3 over F 2
 International Journal of Algebra, Vol. 8, 24, no. 5, 239-246 HIKARI Ltd, www.m-hikari.com http://dx.doi.org/.2988/ija.24.422 Congruence Classes of Invertible Matrices of Order 3 over F 2 Ligong An and
International Journal of Algebra, Vol. 8, 24, no. 5, 239-246 HIKARI Ltd, www.m-hikari.com http://dx.doi.org/.2988/ija.24.422 Congruence Classes of Invertible Matrices of Order 3 over F 2 Ligong An and
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 6/5/2006
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα είναι μικρότεροι το 1000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Διάρκεια: 3,5 ώρες Καλή
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα είναι μικρότεροι το 1000 εκτός αν ορίζεται διαφορετικά στη διατύπωση του προβλήματος. Διάρκεια: 3,5 ώρες Καλή
Review Test 3. MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Review Test MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. Find the exact value of the expression. 1) sin - 11π 1 1) + - + - - ) sin 11π 1 ) ( -
Review Test MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. Find the exact value of the expression. 1) sin - 11π 1 1) + - + - - ) sin 11π 1 ) ( -
