Supplementary Methods
|
|
|
- Ἀελλώ Σαμαράς
- 7 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human ANXA2 rabbit polyclonal antibody (Abcam, Cambridge, UK; dilution 1:1000) and anti-human MSH2 mouse monoclonal antibody (FE11, Dako, Carpinteria, CA; dilution 1:200). Before incubation, the sections were heated for 10 min at 120 o C in an autoclave using sodium citrate buffer at ph 6 and ph 9 (Dako), respectively, for antigen retrieval. Non-specific reactions were blocked with Antibody Diluent (Dako). Both primary antibody incubations were conducted at 4 o C overnight, followed by incubation with EnVision+ Dual link system-hrp (Dako) at room temperature for 30 min. 3.3'-Diaminobenzidine tetrahydrochloride was used as the chromogen. All sections were counterstained with hematoxylin. As a negative control, the primary antibodies were omitted from the reaction sequence. Tissue specimens of T and N samples showing the highest levels of ANXA2 and MSH2 protein expression by the 2DICAL method served as positive controls in all analyses, respectively.
2 VHL (Ch 3, ) Case A57 VHL (Ch 3, ) Case B1 VHL (Ch 3, ) Case A52 VHL (CH 3, ) Case B3 N SNV: A>G SNV: T>A InserKon: T DeleKon: A T PBRM1 (Ch 3, ) Case A42 TNN (Ch 2, ) Case A43 KDM5C (Ch X, ) Case A85 KDM5C (Ch X, ) Case A58 N DeleKon: TT DeleKon: A SNV: C>T DeleKon: T T BAP1 (Ch 3, ) Case A73 BAP1 (Ch 3, ) Case A61 SETD2 (Ch 3, ) Case A69 SETD2 (Ch 3, ) Case B1 N SNV: C>A DeleKon: TT DeleKon: TT DeleKon: GCA T WDFY3 (Ch 4, ) Case A42 WDFY3 (Ch 4, ) Case A50 NCOA1 (Ch 2, ) Case A43 PLCE1 (Ch 10, ) Case B8 N SNV: A>G InserKon: G SNV: C>T SNV: A>T T PARP8 (Ch 5, ) Case B4 CD6 (Ch 11, ) Case B2 CELSR2 (Ch 1, ) TNFSF11 (Ch 13, ) Case B4 Case B3 N SNV: A>G SNV: C>T SNV: C>T DeleKon: A T Supplementary Figure S1. VerificaKon of representakve genekc aberrakons described in Table 2 by Sanger sequencing. RepresentaKve electrophoretogram for Sanger sequencing of the VHL, PBRM1, TNN, KDM5C, BAP1, SETD2, WDFY3, NCOA1, PLCE1, PARP8, CD6, CELSR2 and TNFSF11 genes. PosiKon numbers of the mutakon start site are those based on the NaKonal Center for Biotechnology InformaKon Database Genome Build 37). Ch, chromosome.
3 A B C D Supplementary Figure S2. Representa)ve photos of immunohistochemical examina)ons for ANXA2 (A and B) and MSH2 (C and D) in N (A and C) and T (B and D) samples. These examina)ons were performed as described in Supplementary Methods. (A) Immunoreac)vity for ANXA2 was localized only at the apical membrane of a few proximal tubules, which are the origin of clear cell RCCs, the distal tubules and glomeruli in N samples. (B) Almost all cancer cells showed strong immunoreac)vity for ANXA2 in the circumferen)al cell membrane and cytoplasm in T samples. (C) Almost all epithelial cells showed strong nuclear immunoreac)vity for MSH2 in N samples. (D) Almost all cancer cells lacked immunoreac)vity for MSH2 in T samples. Thus, elevated expression of ANXA2 protein and reduced expression of MSH2 protein were verified immunohistochemically in T samples rela)ve to N samples, indica)ng the reliability of two- dimensional image- converted liquid chromatography- mass spectrometry analysis.
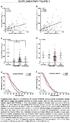
4 mrna expression level (/GAPDH) AURKA P=4.225x P=2.100x P=9.448x10-4 CIMP (-) CIMP (+) AURKB CIMP (-) CIMP (+) BIRC5 CIMP (-) CIMP (+) 8 BUB1 CDC20 NEK2 SPC25 P=5.120x P=1.668x P=5.052x P=5.891x10-3 mrna expression level (/GAPDH) CIMP (-) CIMP (+) CIMP (-) CIMP (+) CIMP (-) CIMP (+) CIMP (-) CIMP (+) Supplementary Figure S3. Levels of mrna expression for genes included in the top pathway Cell cycle: The metaphase checkpoint in CpG island methylator phenotype (CIMP)- negasve renal cell carcinomas (RCCs) (n=74) and CIMP- posisve RCCs (n=19 in the inisal and validason cohorts) evaluated by quanstasve reverse transcripson- PCR analysis. Average levels of mrna expression for AURKA, AURKB, BIRC5, BUB1, CDC20, NEK2 and SPC25 in CIMP- posisve RCCs were significantly higher than those in CIMP- negasve RCCs (Mann- Whitney U test). -, CIMP- negasve RCCs; +, CIMP- posisve RCCs. Error bars, standard error. P values of <0.05 are underlined.
5 Arai E, et al. Supplementary Table S1. Probe and primer sets used for TaqMan Gene Expression Assays for quantitative RT-PCR Gene AURKA AURKB AURKC BIRC5 BUB1 CDC20 NEK2 SPC25 GAPDH Probe and primer set Hs _m1 (Life Technologies) Hs _g1 (Life Technologies) Hs _g1 (Life Technologies) Hs _s1 (Life Technologies) Hs _m1 (Life Technologies) Hs _mH (Life Technologies) Hs _mH (Life Technologies) Hs _m1 (Life Technologies) Hs _g1 (Life Technologies)
6 Supplementary Table S2. All detected genetic aberrations (non-synonymous single nucleotide mutations [SNVs] and insertions and deletions [indels]) in each renal cell carcinoma (RCC) SNVs Gene KLHL17 chr T C exonic NM_198317:c.T284C:p.V95A AGRN chr G A exonic NM_198576:c.G3487A:p.G1163R AGRN chr G A exonic NM_198576:c.G5113A:p.D1705N B3GALT6 chr C T exonic NM_080605:c.C503T:p.A168V MXRA8 chr C G exonic NM_032348:c.G1023C:p.Q341H SLC35E2B chr G T exonic NM_ :c.C689A:p.S230Y PEX10 chr C A exonic NM_002617:c.G381T:p.L127F PLCH2 chr C T exonic NM_014638:c.C464T:p.A155V MMEL1 chr C T exonic stopgain SNV NM_033467:c.G2168A:p.W723X PRDM16 chr A G exonic NM_022114:c.A707G:p.D236G PRDM16 chr G T exonic NM_022114:c.G2953T:p.D985Y TP73 chr A G exonic NM_ :c.A115G:p.N39D LRRC47 chr A C exonic NM_020710:c.T1099G:p.C367G C1orf174 chr G C exonic NM_207356:c.C641G:p.S214C AJAP1 chr G C exonic NM_ :c.G862C:p.V288L ICMT chr T C exonic NM_012405:c.A766G:p.I256V ESPN chr G A exonic NM_031475:c.G974A:p.R325H NOL9 chr T C exonic NM_024654:c.A1262G:p.D421G DNAJC11 chr G A exonic stopgain SNV NM_018198:c.C931T:p.Q311X PER3 chr C A exonic stopgain SNV NM_016831:c.C1269A:p.Y423X PER3 chr C T exonic stopgain SNV NM_016831:c.C2083T:p.R695X SLC2A5 chr C T exonic stopgain SNV NM_003039:c.G720A:p.W240X H6PD chr A T exonic NM_004285:c.A368T:p.D123V KIF1B chr T A exonic NM_015074:c.T4894A:p.L1632M CASZ1 chr G T exonic NM_ :c.C4592A:p.T1531K C1orf127 chr C G exonic NM_ :c.G1877C:p.R626P MTOR chr A C exonic NM_004958:c.T7280G:p.L2427R MTOR chr C T exonic NM_004958:c.G7255A:p.E2419K MTOR chr G T exonic NM_004958:c.C6422A:p.P2141Q MTOR chr A G exonic NM_004958:c.T4447C:p.C1483R MTOR chr G A exonic NM_004958:c.C4360T:p.H1454Y MTOR chr T G exonic NM_004958:c.A4356C:p.K1452N ANGPTL7 chr T A exonic NM_021146:c.T662A:p.V221E ANGPTL7 chr C T exonic NM_021146:c.C946T:p.H316Y PTCHD2 chr C T exonic NM_020780:c.C403T:p.R135W PTCHD2 chr C A exonic NM_020780:c.C4154A:p.P1385H MIIP chr G C exonic NM_021933:c.G46C:p.E16Q VPS13D chr G T exonic NM_015378:c.G981T:p.L327F VPS13D chr T C exonic NM_015378:c.T4925C:p.L1642P FHAD1 chr T A exonic NM_052929:c.T4014A:p.D1338E DNAJC16 chr A T exonic NM_015291:c.A201T:p.E67D AGMAT chr T A exonic NM_024758:c.A442T:p.I148F C1orf64 chr T C exonic stoploss SNV NM_178840:c.T508C:p.X170Q ATP13A2 chr G A exonic NM_ :c.C977T:p.A326V PADI2 chr G A exonic NM_007365:c.C1816T:p.P606S PAX7 chr A T exonic NM_ :c.A673T:p.T225S UBR4(NM_020765:exon80 :c g>t) chr C A sp UBR4 chr T A exonic stopgain SNV NM_020765:c.A1813T:p.K605X KIAA0090 chr G A exonic NM_015047:c.C671T:p.S224F PQLC2 chr C G exonic NM_ :c.C732G:p.S244R EIF4G3 chr C T exonic NM_003760:c.G3436A:p.A1146T EIF4G3 chr T C exonic NM_003760:c.A2999G:p.H1000R ECE1 chr T C exonic NM_ :c.A1553G:p.Y518C CDC42 chr G A exonic NM_001791:c.G17A:p.C6Y ZBTB40 chr A C exonic NM_014870:c.A1406C:p.Y469S ZBTB40 chr C T exonic NM_014870:c.C3101T:p.A1034V EPHA8 chr A T exonic stopgain SNV NM_ :c.A604T:p.K202X EPHB2 chr C T exonic NM_004442:c.C1301T:p.A434V ZNF436 chr T C exonic NM_030634:c.A875G:p.D292G ASAP3 chr T A exonic stoploss SNV NM_ :c.A2684T:p.X895L E2F2 chr G A exonic NM_004091:c.C502T:p.R168C TCEB3 chr G C exonic NM_003198:c.G480C:p.R160S HMGCL chr T G exonic NM_000191:c.A484C:p.I162L GRHL3 chr C A exonic NM_ :c.C976A:p.L326M MAN1C1 chr G A exonic NM_020379:c.G293A:p.S98N SH3BGRL3 chr G T exonic NM_031286:c.G140T:p.R47M UBXN11 chr A T exonic NM_ :c.T1096A:p.C366S UBXN11 chr C G exonic NM_145345:c.G169C:p.V57L ARID1A chr T A exonic stopgain SNV NM_006015:c.T4524A:p.Y1508X MAP3K6 chr G T exonic NM_004672:c.C3632A:p.P1211H SESN2 chr G T exonic NM_031459:c.G821T:p.R274L SRSF4 chr C A exonic NM_005626:c.G767T:p.R256L PTPRU chr C T exonic NM_ :c.C4003T:p.R1335W PEF1 chr C T exonic NM_012392:c.G379A:p.D127N COL16A1 chr C T exonic NM_001856:c.G2522A:p.S841N COL16A1(NM_001856:exo n32:c g>t) chr C A sp TSSK3 chr T G exonic NM_052841:c.T494G:p.L165R S100PBP chr T A exonic NM_ :c.T609A:p.N203K FNDC5;FNDC5 chr T C NM_ :c.A307G:p.I103V TRIM62 chr G T exonic NM_018207:c.C939A:p.D313E PHC2 chr T C exonic NM_004427:c.A275G:p.Y92C CSMD2 chr C T exonic NM_052896:c.G5947A:p.G1983S CSMD2 chr C T exonic NM_052896:c.G3481A:p.A1161T C1orf216 chr T C exonic NM_152374:c.A512G:p.H171R CLSPN chr T C exonic NM_ :c.A2087G:p.D696G EIF2C1 chr C G exonic NM_012199:c.C1220G:p.T407R THRAP3 chr G A exonic NM_005119:c.G1375A:p.A459T OSCP1 chr G T exonic NM_145047:c.C188A:p.S63Y MYCBP chr C G exonic NM_012333:c.G202C:p.E68Q AKIRIN1 chr T A exonic NM_024595:c.T401A:p.L134H MACF1 chr A G exonic NM_012090:c.A1310G:p.Q437R MACF1 chr C A exonic NM_012090:c.C3473A:p.T1158K PABPC4 chr C T exonic NM_ :c.G268A:p.D90N NT5C1A chr G A exonic NM_032526:c.C1079T:p.P360L NT5C1A chr T A exonic NM_032526:c.A214T:p.T72S TRIT1 chr T A exonic NM_017646:c.A430T:p.T144S ZMYND12 chr C G exonic NM_032257:c.G133C:p.D45H YBX1 chr C T exonic NM_004559:c.C749T:p.P250L YBX1 chr A T exonic NM_004559:c.A806T:p.D269V SZT2 chr G A exonic NM_015284:c.G1829A:p.C610Y SZT2 chr C T exonic NM_015284:c.C6164T:p.A2055V IPO13 chr G A exonic NM_014652:c.G405A:p.M135I B4GALT2 chr G A exonic stopgain SNV NM_ :c.G833A:p.W278X BEST4 chr G A exonic stopgain SNV NM_153274:c.C970T:p.Q324X PTCH2 chr C G exonic NM_ :c.G2560C:p.E854Q PTCH2 chr C A exonic stopgain SNV NM_ :c.G2212T:p.E738X EIF2B3 chr C A exonic NM_ :c.G1030T:p.A344S HECTD3 chr C T exonic NM_024602:c.G340A:p.G114S GPBP1L1 chr G T exonic NM_021639:c.C1332A:p.F444L TSPAN1 chr T A exonic NM_005727:c.T707A:p.L236Q C1orf190 chr A G exonic NM_ :c.A557G:p.D186G ATPAF1 chr C T exonic NM_ :c.G469A:p.V157M KIAA0494 chr C T exonic NM_014774:c.G53A:p.R18Q AGBL4 chr T A exonic NM_032785:c.A1380T:p.E460D ELAVL4 chr C G exonic NM_021952:c.C797G:p.P266R ZCCHC11 chr G T exonic NM_ :c.C1917A:p.F639L FAM159A chr G C exonic NM_ :c.G538C:p.V180L ECHDC2 chr C G exonic NM_ :c.G303C:p.Q101H GLIS1 chr A G exonic NM_147193:c.T752C:p.I251T SSBP3 chr C A exonic NM_ :c.G519T:p.M173I HEATR8 chr C G exonic NM_ :c.C248G:p.P83R HEATR8 chr G T exonic NM_ :c.G3951T:p.W1317C TTC4 chr G C exonic NM_004623:c.G654C:p.Q218H TMEM61 chr C T exonic NM_182532:c.C464T:p.A155V PCSK9 chr G A exonic NM_174936:c.G2020A:p.A674T TM2D1 chr C T exonic NM_032027:c.G581A:p.S194N INADL chr A G exonic NM_176877:c.A1780G:p.T594A CACHD1 chr A G exonic NM_020925:c.A1058G:p.N353S RAVER2 chr G C exonic NM_018211:c.G1885C:p.A629P DNAJC6 chr G A exonic NM_014787:c.G755A:p.R252H MIER1 chr A C exonic NM_ :c.A790C:p.N264H WLS chr C T exonic NM_ :c.G8A:p.G3E DEPDC1 chr A C exonic NM_ :c.T1080G:p.D360E LRRIQ3 chr G T exonic NM_ :c.C1367A:p.A456D LRRIQ3 chr T A exonic NM_ :c.A1188T:p.K396N C1orf173 chr T C exonic NM_ :c.A4231G:p.S1411G C1orf173 chr C A exonic NM_ :c.G2022T:p.E674D C1orf173 chr A G exonic NM_ :c.T1511C:p.V504A CRYZ chr A G exonic NM_ :c.T386C:p.I129T CRYZ chr T C exonic NM_ :c.A158G:p.Y53C SLC44A5(NM_ : exon5:c.53-1g>t,nm_152697:exon5:c.53-1g>t) Chromosome Reference sequence Altered sequence chr C A sp Functions Amino acid changes ASB17 chr A G exonic NM_080868:c.T500C:p.L167P ST6GALNAC3 chr A G exonic NM_152996:c.A709G:p.M237V PIGK chr T C exonic NM_005482:c.A802G:p.M268V ZZZ3 chr C T exonic NM_015534:c.G2189A:p.S730N ZZZ3 chr G A exonic NM_015534:c.C1163T:p.T388I DNASE2B chr C T exonic stopgain SNV NM_058248:c.C184T:p.R62X LPAR3 chr T A exonic NM_012152:c.A180T:p.K60N SYDE2 chr G T exonic NM_032184:c.C3140A:p.S1047Y BCL10 chr C A exonic NM_003921:c.G616T:p.D206Y CLCA1 chr C T exonic NM_001285:c.C2612T:p.P871L LRRC8D chr G A exonic NM_ :c.G1083A:p.M361I ZNF326 chr G A exonic NM_182975:c.G147A:p.M49I ZNF644 chr G A exonic stopgain SNV NM_201269:c.C997T:p.Q333X ZNF644 chr A G exonic NM_201269:c.T695C:p.L232S HFM1 chr C T exonic NM_ :c.G3581A:p.R1194H TGFBR3 chr T A exonic NM_ :c.A1778T:p.N593I BRDT chr A T exonic NM_ :c.A805T:p.N269Y GLMN chr T C exonic NM_053274:c.A244G:p.K82E GFI1 chr C T exonic NM_ :c.G1120A:p.G374S BCAR3 chr G T exonic NM_003567:c.C1615A:p.R539S ARHGAP29 chr G T exonic NM_004815:c.C2591A:p.S864Y ABCD3 chr A C exonic NM_002858:c.A1214C:p.K405T PTBP2 chr A G exonic NM_021190:c.A892G:p.T298A DPYD chr T A exonic NM_000110:c.A2516T:p.Y839F FRRS1 chr G A exonic NM_ :c.C1175T:p.A392V VCAM1 chr T A exonic NM_080682:c.T1868A:p.M623K COL11A1 chr G A exonic NM_080630:c.C4949T:p.T1650I COL11A1 chr G T exonic NM_080630:c.C1681A:p.P561T RNPC3 chr G C exonic NM_017619:c.G1377C:p.L459F PRMT6 chr G C exonic NM_018137:c.G364C:p.E122Q FAM102B chr G T exonic NM_ :c.G61T:p.V21L STXBP3 chr T G exonic stopgain SNV NM_007269:c.T1469G:p.L490X AKNAD1 chr A T exonic NM_152763:c.T2488A:p.W830R WDR47 chr T C exonic NM_ :c.A2140G:p.M714V CELSR2 chr C T exonic NM_001408:c.C8393T:p.A2798V CELSR2 chr G A exonic NM_001408:c.G8435A:p.R2812Q MYBPHL chr G A exonic NM_ :c.C797T:p.P266L SORT1 chr G C exonic NM_ :c.C993G:p.I331M PSMA5 chr C A exonic NM_ :c.G295T:p.D99Y Case ID (average coverage for tumorous tissue [T] sample) "Non-reference read number/total (non-reference and reference) read number in T sample; non-reference read number/total read number in N sample" for each genetic aberration has been shown. A1 (143.3 ) A2 (106.6 ) 12/31; 0/24 A3 (147.4 ) 37/251 ;0/247 31/174 ;0/166 B1 (153.9 ) 25/192 ;0/141 31/230 ;1/164 A4 (136.7 ) 43/146 ;0/126 20/52; 1/61 51/197 ;1/173 26/123 ;0/116 A5 (142.4 ) 35/108 ;0/105 B2 (102.4 ) 30/67; 0/59 7/52;0/ 78 A6 (131.0 ) B3 (114.4 ) 18/108 ;1/150 B4 (125.8 ) 36/92; 1/82 63/162 ;2/151 27/64; 0/55 A7 (102.1 ) 21/130 ;1/111 A8 (137.7 ) 25/70; 0/46 8/60;0/ 42 A9 (123.9 ) A10 (121.7 ) 24/96; 0/64 16/83; 0/81 A11 (112.6 ) 9/59;0/ 53 A12 (100.5 ) 18/57; 0/44 A13 (118.0 ) 24/73; 0/68 91/381 ;0/372 A14 (110.4 ) 20/61; 0/56 4/31;0/ 25 28/99; 0/124 A15 (101.9 ) 21/85; 0/124 A16 (113.4 ) A17 (128.7 ) 11/46; 0/54 A18 (110.0 ) 18/64; 0/69 A19 (96.2 ) 39/122 ;1/134 A20 (120.7 ) 34/143 ;1/104 12/88; 0/92 45/232 ;1/233 A21 (126.6 ) 15/42; 0/37 33/106 ;0/120 28/106 ;0/107 A22 (101.8 ) 21/72; 1/63 47/148 ;0/150 B5 (114.8 ) 19/82; 0/73 13/113 ;0/122 2/15;0/ 21 11/103 ;0/99 B6 (119.0 ) 19/127 ;2/150 30/181 ;2/226 12/94; 1/127 8/49;0/ 55 A23 (107.2 ) 53/171 ;0/191 9/71;0/ 80 35/132 ;0/124 A24 (125.1 ) 4/18;0/ 16 A25 (115.3 ) A26 (133.9 ) 4/22;0/ 20 18/82; 0/104 34/145 ;0/152 A27 (138.7 ) 18/106 ;0/71 22/107 ;0/88 48/368 ;1/375 A28 (150.8 ) A29 (114.0 ) 9/85;1/ 87 A30 (123.9 ) 25/181 ;0/163 A31 (107.0 ) 8/57;0/ 86 35/153 ;0/181 4/30;0/ 44 A32 (129.8 ) A33 (110.8 ) 28/118 ;0/105 A34 (125.3 ) 56/204 ;1/210 22/69; 1/73 16/78; 0/53 97/333 ;2/335 B7 (115.9 ) 4/26;0/ 24 14/84; 0/86 8/63;0/ 58 16/116 ;1/121 10/92; 2/105 8/42;0/ 54 9/50;0/ 55 11/86; 0/97 15/129 ;0/159 9/62;1/ 81 8/65;0/ 83 9/80;0/ 73 14/122 ;1/117 4/28;0/ 45 A35 (110.7 ) 3/23;0/ 31 11/93; 0/93 18/78; 1/98 32/114 ;1/133 B8 (103.4 ) 8/48;0/ 74 67/210 ;0/266 56/211 ;2/257 A36 (125.4 ) A37 (123.6 ) 23/82; 0/101 A38 (119.2 ) 2/18;0/ 25 A39 (108.5 ) A40 (141.1 ) 18/60; 1/54 25/56; 1/96 17/47; 1/61 43/186 ;0/ /72 7;1/ /145 ;1/187 B9 (150.4 ) 30/124 ;0/110 16/98; 0/112 22/203 ;0/160 A41 (116.8 ) 14/67; 1/94 15/90; 0/75 A42 (147.4 ) 8/23;0/ 32 5/25;0/ 20 43/174 ;1/153 17/113 ;0/87 70/367 ;0/357 A43 (141.9 ) 25/55; 1/78 18/88; 0/ /26 3;1/ /26 7;2/25 7 A44 (124.3 ) 15/46; 0/74 10/54; 0/56 A45 (132.3 ) A46 (128.6 ) 16/84; 0/75 13/66; 0/73 21/89; 0/110 A47 (137.9 ) 61/169 ;2/214 B10 (139.2 ) 11/59; 0/53 58/231 ;2/257 A48 (133.6 ) A49 (131.1 ) A50 (96.0 ) A51 (127.4 ) A52 (126.5 ) 28/131 ;0/120 12/45; 0/51 A53 (99.2 ) B11 (98.4 ) 31/67; 0/108 A54 (144.2 ) A55 (144.9 ) A56 (147.8 ) 20/117 ;0/112 21/110 ;0/102 A57 (148.6 ) 5/46;0/ 23 2/18;0/ 16 13/115 ;0/105 53/208 ;0/159 53/224 ;2/229 76/314 ;1/287 A58 (132.6 ) 24/103 ;0/125 A59 (109.4 ) 11/23; 0/27 47/191 ;0/199 A60 (104.3 ) A61 (142.5 ) 42/191 ;0/183 20/143 ;0/147 A62 (100.2 ) 39/138 ;0/155 A63 (132.7 ) A64 (137.5 ) 8/63;0/ 54 A65 (148.4 ) 8/49;0/ 54 A66 (135.8 ) 5/44;1/ 51 52/176 ;0/200 A67 (136.3 ) 58/196 ;0/157 A68 (128.3 ) 21/77; 0/88 35/299 ;0/384 2/15;0/ 21 2/15;0/ 16 A69 (134.9 ) 17/137 ;1/164 A70 (100.1 ) A71 (145.2 ) 2/10;0/ 19 22/48; 0/49 A72 (140.2 ) 30/177 ;0/214 A73 (147.1 ) 2/15;0/ 20 A74 (156.5 ) 2/10;0/ 16 28/187 ;1/197 7/50;0/ 62 39/269 ;0/241 A75 (103.3 ) 14/102 ;0/116 A76 (91.2 ) 7/59;0/ 71 10/82; 0/94 3/22;0/ 40 A77 (94.7 ) A78 (106.9 ) 51/289 ;0/288 80/226 ;0/214 A79 (139.4 ) 58/251 ;0/198 49/222 ;1/207 A80 (102.4 ) 17/76; 0/70 17/52; 0/51 B12 (115.5 ) A81 (137.1 ) 2/18;0/ 24 B13 (105.5 ) 16/92; 0/92 14/106 ;0/105 A82 (109.0 ) 33/170 ;3/205 9/41;0/ 23 39/200 ;0/181 A83 (109.1 ) 26/53; 0/78 2/13;0/ 23 B14 (111.2 ) 30/103 ;0/81 32/108 ;0/113 A84 (128.1 ) 19/163 ;0/156 A85 (134.0 ) A86 (144.3 ) 13/33; 0/23 17/59; 1/55 21/61; 0/51 A87 (146.4 ) 13/56; 0/63 12/69; 0/77 37/250 ;0/241 Start position * End position *
7 EPS8L3 chr G T exonic NM_024526:c.C1582A:p.Q528K EPS8L3 chr A T exonic NM_024526:c.T927A:p.S309R SLC16A4 chr T A exonic NM_ :c.A121T:p.I41F LRIF1 chr G T exonic NM_018372:c.C38A:p.A13E CEPT1 chr A T exonic NM_ :c.A11T:p.H4L AP4B1(NM_006594:exon3: c.113+2t>c) chr A G sp DCLRE1B chr G C exonic NM_022836:c.G559C:p.E187Q HIPK1 chr C G exonic stopgain SNV NM_181358:c.C120G:p.Y40X OLFML3 chr A C exonic NM_020190:c.A843C:p.E281D TRIM33 chr A G exonic NM_015906:c.T2801C:p.I934T SYCP1;SYCP1 chr G C NM_003176:c.G802C:p.V268L TSHB chr C A exonic NM_000549:c.C28A:p.L10I MAB21L3 chr C T exonic stopgain SNV NM_152367:c.C637T:p.Q213X MAB21L3 chr T G exonic stoploss SNV NM_152367:c.T1087G:p.X363G PTGFRN chr A G exonic NM_020440:c.A1496G:p.N499S SPAG17 chr C A exonic NM_206996:c.G4790T:p.G1597V HMGCS2 chr C G exonic NM_ :c.G1391C:p.R464P HMGCS2 chr C T exonic stopgain SNV NM_ :c.G362A:p.W121X NOTCH2 chr C T exonic NM_024408:c.G4800A:p.M1600I POLR3GL chr A G exonic NM_032305:c.T614C:p.F205S ITGA10 chr C T exonic NM_003637:c.C2132T:p.A711V ITGA10 chr G A exonic NM_003637:c.G2465A:p.R822Q POLR3C chr T C exonic NM_006468:c.A1420G:p.M474V HIST2H2AB chr T C exonic NM_175065:c.A229G:p.T77A SV2A chr C T exonic NM_014849:c.G1858A:p.D620N SV2A chr T A exonic NM_014849:c.A431T:p.E144V MTMR11 chr C T exonic NM_181873:c.G280A:p.A94T VPS45 chr G T exonic NM_007259:c.G451T:p.D151Y TUFT1 chr C T exonic NM_ :c.C808T:p.L270F TCHH chr G A exonic NM_007113:c.C5626T:p.R1876W TCHH chr C T exonic NM_007113:c.G5308A:p.D1770N HRNR chr C T exonic NM_ :c.G1067A:p.G356E FLG chr G A exonic NM_002016:c.C3706T:p.R1236C FLG2 chr C A exonic NM_ :c.G2783T:p.G928V SPRR4 chr C A exonic NM_173080:c.C58A:p.Q20K NUP210L chr C T exonic NM_ :c.G839A:p.G280E NUP210L chr C T exonic NM_ :c.G838A:p.G280R UBAP2L chr T A exonic stopgain SNV NM_ :c.T642A:p.Y214X ADAR chr C T exonic NM_ :c.G1411A:p.G471S ZBTB7B chr A T exonic NM_015872:c.A1511T:p.Q504L ADAM15 chr G A exonic NM_003815:c.G100A:p.A34T SLC50A1 chr C A exonic NM_ :c.C47A:p.P16H SLC50A1 chr T C exonic NM_ :c.T248C:p.I83T FDPS chr G A exonic NM_ :c.G265A:p.G89S FDPS chr G A exonic NM_ :c.G685A:p.A229T RUSC1 chr C A exonic NM_ :c.C482A:p.S161Y GON4L chr G T exonic NM_ :c.C805A:p.L269M KIAA0907 chr C T exonic NM_014949:c.G1696A:p.A566T UBQLN4 chr A C exonic NM_020131:c.T1339G:p.F447V TMEM79 chr G T exonic NM_032323:c.G284T:p.R95L TMEM79 chr C G exonic stopgain SNV NM_032323:c.C1074G:p.Y358X GPATCH4 chr C G exonic NM_015590:c.G975C:p.R325S GPATCH4 chr C T exonic NM_015590:c.G824A:p.G275E PEAR1 chr C T exonic NM_ :c.C1444T:p.P482S ARHGEF11 chr C T exonic NM_014784:c.G1735A:p.D579N ARHGEF11(NM_198236:e xon3:c.33-2a>c,nm_014784:exon3:c.33-2a>c) chr T G sp FCRL5 chr T C exonic NM_ :c.A718G:p.S240G FCRL2 chr A G exonic NM_030764:c.T1102C:p.C368R SPTA1 chr C G exonic NM_003126:c.G5692C:p.E1898Q SPTA1(NM_003126:exon3 9:c T>C) chr A G sp SPTA1 chr A T exonic NM_003126:c.T2594A:p.F865Y SPTA1 chr T A exonic NM_003126:c.A322T:p.M108L OR6K2 chr A C exonic NM_ :c.T446G:p.V149G OR6K3 chr G A exonic NM_ :c.C284T:p.T95I PYHIN1 chr C A exonic NM_152501:c.C1315A:p.P439T CCDC19 chr T C exonic NM_012337:c.A1292G:p.K431R IGSF9 chr A G exonic NM_ :c.T1504C:p.Y502H IGSF8 chr T G exonic NM_052868:c.A1819C:p.M607L DCAF8 chr A G exonic NM_015726:c.T1451C:p.L484P SLAMF6 chr G A exonic NM_ :c.C239T:p.P80L ARHGAP30 chr T A exonic NM_ :c.A1045T:p.S349C B4GALT3 chr G A exonic NM_ :c.C1123T:p.P375S ATF6 chr G C exonic NM_007348:c.G1915C:p.D639H OLFML2B chr C T exonic NM_015441:c.G863A:p.R288Q NOS1AP chr A G exonic NM_ :c.A326G:p.Y109C GPR161 chr G A exonic NM_153832:c.C908T:p.P303L SELL chr G A exonic NM_000655:c.C755T:p.T252I C1orf112 chr A T exonic NM_018186:c.A1571T:p.E524V C1orf112 chr A C exonic NM_018186:c.A1577C:p.E526A C1orf112 chr C A exonic NM_018186:c.C1784A:p.T595K GORAB chr C T exonic NM_ :c.C337T:p.H113Y C1orf129 chr T A exonic NM_ :c.T1558A:p.S520T METTL13 chr G A exonic NM_014955:c.G544A:p.V182M PIGC chr C G exonic NM_002642:c.G567C:p.L189F RC3H1 chr T C exonic NM_172071:c.A1913G:p.Y638C TNN chr G A exonic NM_022093:c.G472A:p.E158K TNR chr C G exonic NM_003285:c.G3613C:p.E1205Q SEC16B chr C T exonic NM_033127:c.G877A:p.V293I TOR3A chr A G exonic NM_022371:c.A445G:p.R149G TOR1AIP1 chr A C exonic NM_015602:c.A707C:p.K236T CEP350 chr G A exonic NM_014810:c.G3611A:p.R1204H CEP350 chr C T exonic NM_014810:c.C5870T:p.P1957L XPR1 chr C T exonic NM_ :c.C1319T:p.S440L SHCBP1L chr G T exonic NM_030933:c.C1072A:p.L358I LAMC2 chr T C exonic NM_005562:c.T202C:p.C68R LAMC2 chr G A exonic NM_005562:c.G2428A:p.G810S TRMT1L chr T C exonic NM_ :c.A1295G:p.E432G SWT1 chr G T exonic NM_ :c.G458T:p.S153I HMCN1 chr G A exonic NM_031935:c.G16784A:p.R5595Q HMCN1 chr A C exonic NM_031935:c.A16797C:p.E5599D TPR chr T G exonic NM_003292:c.A2442C:p.L814F PTGS2 chr C A exonic NM_000963:c.G738T:p.E246D PTGS2 chr C T exonic NM_000963:c.G484A:p.E162K PLA2G4A chr C T exonic stopgain SNV NM_024420:c.C598T:p.R200X FAM5C chr A T exonic NM_199051:c.T1100A:p.L367Q UCHL5 chr A G exonic NM_ :c.T287C:p.V96A KCNT2 chr A C exonic NM_198503:c.T1897G:p.L633V CFH chr G A exonic NM_000186:c.G1708A:p.E570K ASPM chr T A exonic NM_ :c.A128T:p.H43L CRB1 chr C T exonic NM_ :c.C1898T:p.T633M CRB1 chr C G exonic NM_ :c.C2457G:p.S819R DENND1B chr G C exonic NM_ :c.C703G:p.L235V ZNF281 chr T C exonic NM_012482:c.A1027G:p.K343E KIF14 chr C T exonic NM_014875:c.G4760A:p.S1587N KIF14 chr T A exonic NM_014875:c.A3863T:p.Q1288L C1orf106 chr T C exonic NM_ :c.T41C:p.I14T IGFN1 chr G A exonic NM_ :c.G7946A:p.R2649K IGFN1 chr C T exonic NM_ :c.C10841T:p.P3614L PKP1 chr G A exonic NM_000299:c.G494A:p.G165D LGR6 chr C T exonic NM_ :c.C203T:p.T68M LGR6 chr T G exonic NM_ :c.T310G:p.Y104D KDM5B chr T C exonic NM_006618:c.A568G:p.S190G MYOG chr G T exonic NM_002479:c.C540A:p.S180R ATP2B4 chr C T exonic NM_ :c.C244T:p.H82Y LAX1 chr C T exonic NM_017773:c.C17T:p.P6L REN chr T A exonic NM_000537:c.A364T:p.T122S PLEKHA6 chr G C exonic NM_014935:c.C540G:p.D180E PLEKHA6 chr G T exonic NM_014935:c.C196A:p.L66I PPP1R15B(NM_032833:ex on3:c a>c) chr T G sp PIK3C2B chr G A exonic NM_002646:c.C401T:p.S134L NUAK2 chr G T exonic NM_030952:c.C1465A:p.P489T SLC26A9 chr C T exonic NM_052934:c.G886A:p.A296T DYRK3 chr A T exonic NM_003582:c.A714T:p.K238N FAIM3 chr T C exonic NM_ :c.A361G:p.N121D CR2 chr A G exonic NM_ :c.A851G:p.N284S CR2 chr A C exonic NM_ :c.A1253C:p.N418T CR2 chr C T exonic NM_ :c.C2030T:p.T677M NEK2 chr C A exonic stopgain SNV NM_ :c.G727T:p.E243X FAM71A chr T C exonic NM_153606:c.T158C:p.I53T FLVCR1 chr T C exonic NM_014053:c.T1244C:p.I415T ANGEL2 chr G A exonic NM_144567:c.C217T:p.H73Y ANGEL2 chr A C exonic NM_144567:c.T206G:p.F69C PROX1 chr G A exonic NM_002763:c.G467A:p.R156H CENPF chr T G exonic NM_016343:c.T8708G:p.L2903R USH2A chr T A exonic NM_206933:c.A10085T:p.E3362V USH2A chr G C exonic NM_206933:c.C6908G:p.S2303C ESRRG chr T G exonic NM_ :c.A564C:p.K188N DUSP10 chr G C exonic NM_144728:c.C357G:p.D119E MIA3 chr C T exonic NM_198551:c.C3508T:p.P1170S CAPN2 chr G A exonic NM_ :c.G130A:p.A44T CAPN2 chr C G exonic NM_ :c.C513G:p.D171E DEGS1 chr C T exonic NM_003676:c.C344T:p.P115L TMEM63A chr G A exonic stopgain SNV NM_014698:c.C2323T:p.Q775X TMEM63A chr C T exonic NM_014698:c.G1700A:p.G567D TMEM63A chr C A exonic NM_014698:c.G510T:p.L170F PARP1 chr C T exonic NM_001618:c.G2633A:p.R878Q ZNF678 chr G T exonic NM_178549:c.G660T:p.W220C ZNF678 chr G C exonic NM_178549:c.G1175C:p.S392T PRSS38(NM_183062:exon 2:c.311+1G>C) chr G C sp OBSCN chr G T exonic NM_ :c.G10179T:p.W3393C OBSCN chr C A exonic NM_ :c.C15171A:p.F5057L URB2 chr G T exonic NM_014777:c.G708T:p.Q236H AGT chr A G exonic NM_000029:c.T551C:p.L184P ARV1 chr A G exonic NM_022786:c.A212G:p.Y71C C1orf124 chr T G exonic NM_032018:c.T868G:p.S290A TSNAX chr T A exonic NM_005999:c.T566A:p.L189Q DISC1 chr C G exonic NM_ :c.C555G:p.N185K SIPA1L2 chr G A exonic NM_020808:c.C4670T:p.S1557F PCNXL2 chr A T exonic NM_014801:c.T2620A:p.C874S PCNXL2 chr T C exonic NM_014801:c.A2168G:p.K723R SLC35F3 chr C T exonic NM_173508:c.C292T:p.R98C TARBP1 chr T C exonic NM_005646:c.A1796G:p.K599R IRF2BP2 chr T C exonic NM_ :c.A1115G:p.K372R ARID4B chr G C exonic NM_031371:c.C2031G:p.N677K LYST chr T A exonic stopgain SNV NM_000081:c.A7270T:p.K2424X LYST chr C G exonic NM_000081:c.G3880C:p.E1294Q NID1 chr C T exonic NM_002508:c.G406A:p.E136K HEATR1 chr A T exonic NM_018072:c.T3412A:p.C1138S 29/93; 0/94 2/19;0/ 32 8/47;0/ 57 22/161 ;0/ /45 7;1/ /55; 0/60 20/71; 1/72 33/108 ;1/127 7/62;1/ 78 10/76; 1/88 22/192 ;0/156 26/93; 1/98 26/113 ;1/144 10/85; 0/118 4/34;0/ 56 36/95; 1/82 33/207 ;1/264 19/134 ;0/142 6/32;0/ 30 27/170 ;1/145 2/18;0/ 25 39/165 ;0/134 34/129 ;1/101 72/304 ;0/318 23/108 ;0/103 20/87; 0/89 51/112 ;0/125 46/96; 0/87 15/96; 0/94 11/57; 0/57 13/47; 0/63 25/88; 0/102 24/106 ;0/103 29/121 ;0/101 47/217 ;0/220 21/91; 0/74 11/65; 0/70 2/11;0/ 18 15/66; 0/58 11/42; 0/39 39/121 ;0/128 9/39;0/ 39 42/148 ;0/187 7/32;0/ 45 30/81; 0/84 17/103 ;0/156 21/55; 0/62 27/72; 0/78 36/113 ;0/125 6/38;0/ 63 26/157 ;0/156 14/106 ;0/92 32/139 ;0/126 25/90; 0/84 10/88; 1/96 12/45; 0/42 50/243 ;0/238 13/82; 0/87 28/112 ;1/91 20/77; 0/78 29/148 ;1/135 22/105 ;0/95 7/32;0/ 18 44/165 ;2/225 2/17;0/ 21 2/16;0/ 17 15/49; 0/50 21/77; 0/103 35/142 ;0/178 35/337 ;3/374 15/143 ;2/131 11/75; 0/94 10/77; 0/91 3/29;0/ 36 33/194 ;0/220 4/12;0/ 16 16/125 ;0/177 13/109 ;0/140 32/285 ;0/319 24/129 ;0/132 2/10;0/ 15 23/81; 0/79 50/130 ;0/114 16/98; 0/106 66/265 ;0/255 43/148 ;0/129 22/204 ;0/220 11/93; 0/82 24/121 ;0/150 14/57; 0/56 41/131 ;1/119 2/18;0/ 15 44/153 ;1/169 23/166 ;0/144 94/215 ;0/197 13/78; 0/96 2/16;0/ 19 14/89; 0/75 45/136 ;0/131 47/236 ;0/206 36/230 ;3/274 48/249 ;0/262 33/141 ;0/120 2/16;0/ 19 51/233 ;2/270 81/163 ;0/117 87/241 ;2/281 12/109 ;0/126 30/127 ;0/124 15/56; 0/54 15/53; 0/55 25/73; 0/58 122/39 7;4/ /82; 0/69 44/135 ;1/136 6/28;0/ 17 55/137 ;1/138 2/9;0/1 6 49/186 ;0/152 46/134 ;0/129 39/151 ;1/142 30/168 ;0/142 16/83; 1/84 51/236 ;0/170 51/236 ;0/170 87/164 ;0/ /51 8;2/ /177 ;0/211 18/83; 0/116 26/112 ;0/107 24/77; 0/96 48/223 ;1/ /36 6;0/ /86; 0/89 75/194 ;1/201 77/204 ;1/211 52/385 ;0/399 10/92; 0/96 68/270 ;0/313 10/67; 0/80 43/130 ;0/130 24/69; 0/58 12/94; 0/100 17/107 ;0/108 13/82; 0/100 15/80; 0/93 21/109 ;1/127 20/78; 0/68 46/178 ;1/196 34/161 ;1/177 24/94; 0/104 20/87; 0/102 29/170 ;0/168 10/55; 0/53 26/117 ;0/99 18/147 ;0/189 6/30;0/ 32 32/146 ;1/133 52/193 ;0/148 7/49;0/ 47 2/13;0/ 17 5/23;0/ 22 89/405 ;3/335 15/113 ;1/127 67/288 ;0/217 21/94; 0/84 23/85; 0/62 56/229 ;1/208 18/79; 0/86 37/243 ;0/206 48/151 ;0/118 33/116 ;0/107 31/135 ;1/110 63/290 ;0/ /46 2;2/47 2 8/62;0/ 58 17/61; 0/66 8/45;0/ 37
8 MTR chr A C exonic NM_000254:c.A664C:p.I222L RYR2 chr G A exonic NM_001035:c.G3997A:p.D1333N RYR2 chr T C exonic NM_001035:c.T4580C:p.L1527P RYR2 chr A C exonic NM_001035:c.A5299C:p.S1767R RYR2 chr A C exonic NM_001035:c.A12428C:p.K4143T RYR2 chr C T exonic NM_001035:c.C13897T:p.L4633F FMN2 chr C G exonic NM_020066:c.C1084G:p.P362A FMN2 chr G T exonic stopgain SNV NM_020066:c.G1738T:p.E580X FMN2 chr C T exonic NM_020066:c.C3427T:p.P1143S FMN2 chr G A exonic NM_020066:c.G4282A:p.A1428T ADSS chr C T exonic NM_001126:c.G1010A:p.R337K ADSS chr T C exonic NM_001126:c.A488G:p.K163R HNRNPU chr T A exonic NM_004501:c.A780T:p.E260D KIF26B chr G T exonic NM_018012:c.G2701T:p.G901C KIF26B chr G A exonic NM_018012:c.G4780A:p.G1594R KIF26B chr G A exonic NM_018012:c.G6071A:p.R2024H ZNF496 chr G T exonic NM_032752:c.C71A:p.P24Q OR2W5 chr G A exonic NM_ :c.G610A:p.G204S OR2G2 chr A C exonic NM_ :c.A15C:p.R5S OR1C1 chr C G exonic NM_012353:c.G149C:p.G50A OR2M3 chr G T exonic NM_ :c.G738T:p.L246F ZNF672 chr C T exonic NM_024836:c.C997T:p.H333Y PGBD2 chr G C exonic NM_170725:c.G166C:p.D56H DIP2C chr C A exonic NM_014974:c.G4555T:p.V1519L AKR1E2 chr A C exonic NM_ :c.A410C:p.E137A FBXO18 chr T G exonic NM_178150:c.T576G:p.I192M IL2RA chr G T exonic NM_000417:c.C574A:p.Q192K PRKCQ chr A G exonic NM_ :c.T536C:p.F179S ITIH2 chr C A exonic NM_002216:c.C1554A:p.F518L TAF3 chr G T exonic NM_031923:c.G2534T:p.R845L UPF2 chr T C exonic NM_015542:c.A3779G:p.N1260S CAMK1D chr C T exonic NM_020397:c.C823T:p.R275W FAM171A1 chr A T exonic NM_ :c.T854A:p.M285K C1QL3 chr G T exonic NM_ :c.C13A:p.L5M CUBN;CUBN chr G A stopgain SNV NM_001081:c.C8755T:p.R2919X CUBN chr A G exonic NM_001081:c.T5716C:p.Y1906H CUBN chr T C exonic NM_001081:c.A3838G:p.N1280D CUBN chr T A exonic NM_001081:c.A2756T:p.H919L CUBN;CUBN chr C A NM_001081:c.G490T:p.G164C CACNB2 chr G T exonic NM_000724:c.G1198T:p.D400Y COMMD3-BMI1 chr A C exonic NM_ :c.A428C:p.E143A PIP4K2A(NM_005028:exo n3:c.145-2a>t) chr T A sp ARMC3 chr T G exonic NM_173081:c.T569G:p.L190R ARMC3 chr C G exonic NM_173081:c.C1142G:p.A381G KIAA1217 chr C A exonic NM_ :c.C407A:p.P136Q KIAA1217 chr G A exonic NM_ :c.G1780A:p.V594M KIAA1217 chr C A exonic NM_ :c.C3038A:p.P1013H KIAA1217 chr C G exonic NM_ :c.C3344G:p.P1115R MYO3A chr A T exonic NM_017433:c.A2662T:p.N888Y MYO3A chr C G exonic NM_017433:c.C3171G:p.D1057E MYO3A chr A G exonic NM_017433:c.A3629G:p.E1210G ANKRD26 chr C T exonic NM_014915:c.G5101A:p.V1701I ANKRD26 chr T G exonic NM_014915:c.A3320C:p.Q1107P ANKRD26 chr T C exonic NM_014915:c.A1705G:p.T569A ACBD5;ACBD5 chr T C NM_ :c.A271G:p.I91V PTCHD3 chr G A exonic NM_ :c.C290T:p.P97L ARMC4 chr T G exonic NM_018076:c.A2261C:p.E754A SVIL chr C T exonic NM_003174:c.G5131A:p.E1711K SVIL chr G A exonic NM_003174:c.C842T:p.S281L LYZL2 chr C T exonic NM_183058:c.G125A:p.R42Q ZNF438 chr C T exonic NM_ :c.G1853A:p.G618D PARD3;PARD3 chr A T NM_ :c.T2239A:p.S747T BMS1 chr G T exonic stopgain SNV NM_014753:c.G1414T:p.E472X OR13A1 chr C T exonic NM_ :c.G376A:p.A126T WDFY4 chr G A exonic NM_020945:c.G6814A:p.G2272S FAM170B chr G A exonic NM_ :c.C145T:p.P49S C10orf71 chr G T exonic stopgain SNV NM_ :c.G3946T:p.E1316X PGBD3 chr A G exonic NM_170753:c.T464C:p.I155T CHAT chr G T exonic NM_ :c.G1358T:p.R453I CHAT chr C A exonic NM_ :c.C1816A:p.L606M BICC1 chr A G exonic NM_ :c.A1826G:p.E609G PHYHIPL chr A T exonic NM_ :c.A962T:p.E321V FAM13C chr C T exonic NM_198215:c.G1135A:p.A379T SLC16A9 chr A G exonic NM_194298:c.T413C:p.L138P SLC16A9 chr C A exonic NM_194298:c.G265T:p.V89L ARID5B chr G A exonic NM_ :c.G2413A:p.E805K JMJD1C chr G A exonic stopgain SNV NM_004241:c.C250T:p.Q84X HERC4 chr C G exonic NM_015601:c.G2005C:p.D669H DNA2 chr C T exonic NM_ :c.G1540A:p.A514T SLC25A16 chr T C exonic NM_152707:c.A790G:p.T264A STOX1 chr A G exonic NM_ :c.A1280G:p.H427R DDX50 chr G C exonic NM_024045:c.G1261C:p.D421H KIAA1279 chr A T exonic stopgain SNV NM_015634:c.A1324T:p.R442X TACR2 chr C T exonic NM_001057:c.G662A:p.G221D KIAA1274 chr G C exonic NM_014431:c.G271C:p.E91Q ASCC1 chr G C exonic NM_ :c.C532G:p.P178A OIT3 chr C A exonic NM_152635:c.C249A:p.H83Q P4HA1;P4HA1(NM_ :exon11:c G>A ) chr C T NM_000917:c.G1249A:p.V417I USP54 chr G C exonic NM_152586:c.C2120G:p.S707W USP54 chr C G exonic NM_152586:c.G693C:p.E231D SYNPO2L chr T C exonic NM_024875:c.A1301G:p.N434S KIAA0913 chr G A exonic NM_ :c.G2854A:p.D952N NDST2 chr G T exonic NM_003635:c.C221A:p.P74H FAM213A chr T A exonic NM_025125:c.T200A:p.L67H ANXA11 chr G A exonic NM_001157:c.C352T:p.P118S DYDC2 chr C A exonic stopgain SNV NM_032372:c.C39A:p.C13X RGR chr C A exonic NM_ :c.C244A:p.P82T FAM190B chr C T exonic NM_018999:c.C298T:p.P100S GRID1 chr C G exonic NM_017551:c.G1645C:p.E549Q WAPAL chr G T exonic NM_015045:c.C1127A:p.T376N MMRN2 chr C G exonic NM_024756:c.G1723C:p.V575L ATAD1(NM_032810:exon5: c.382+2t>a) chr A T sp PTEN chr G T exonic stopgain SNV NM_000314:c.G118T:p.E40X PTEN chr C A exonic NM_000314:c.C511A:p.Q171K LIPN chr T G exonic NM_ :c.T213G:p.H71Q LIPM chr A T exonic NM_ :c.A262T:p.T88S IFIT3 chr T C exonic NM_ :c.T2C:p.M1T PCGF5 chr C T exonic NM_032373:c.C95T:p.T32M PCGF5 chr G A exonic NM_032373:c.G500A:p.C167Y BTAF1 chr A G exonic NM_003972:c.A1747G:p.I583V EXOC6 chr G T exonic NM_ :c.G2130T:p.Q710H LGI1 chr A C exonic NM_005097:c.A1112C:p.Q371P PLCE1 chr G C exonic NM_ :c.G148C:p.E50Q PLCE1 chr G A exonic NM_ :c.G1971A:p.M657I PLCE1 chr G T exonic NM_ :c.G4366T:p.A1456S PLCE1 chr A C exonic NM_ :c.A4400C:p.K1467T PLCE1 chr A T exonic NM_ :c.A4401T:p.K1467N NOC3L chr A G exonic NM_022451:c.T158C:p.V53A TBC1D12 chr A C exonic NM_015188:c.A2175C:p.K725N DNTT chr C A exonic NM_ :c.C427A:p.P143T LCOR chr T C exonic NM_ :c.T583C:p.W195R MMS19 chr G C exonic NM_022362:c.C1769G:p.A590G ZFYVE27 chr C T exonic NM_ :c.C86T:p.P29L PYROXD2 chr G C exonic NM_032709:c.C1097G:p.S366C HPS1 chr G A exonic NM_000195:c.C1652T:p.T551I ABCC2 chr A G exonic NM_000392:c.A1783G:p.M595V ABCC2(NM_000392:exon2 4:c G>A) chr G A sp DNMBP chr A G exonic NM_015221:c.T602C:p.L201P SEMA4G;SEMA4G chr A T NM_ :c.A812T:p.K271M SFXN3 chr C T exonic stopgain SNV NM_030971:c.C922T:p.Q308X PPRC1 chr C T exonic stopgain SNV NM_015062:c.C4681T:p.Q1561X SUFU;SUFU chr G T NM_ :c.G182T:p.W61L TAF5 chr G A exonic NM_006951:c.G2193A:p.M731I CALHM3 chr G T exonic NM_ :c.C738A:p.F246L CCDC147 chr G C exonic NM_ :c.G804C:p.E268D XPNPEP1 chr T G exonic NM_ :c.A1345C:p.T449P XPNPEP1 chr T G exonic NM_ :c.A1191C:p.K397N ADD3 chr G T exonic NM_001121:c.G1061T:p.G354V MXI1 chr A G exonic NM_ :c.A442G:p.I148V ADRA2A chr C A exonic stopgain SNV NM_000681:c.C420A:p.C140X NRAP(NM_198060:exon43 :c a>t,nm_006175:exon42: c a>t) chr T A sp NRAP chr G A exonic NM_006175:c.C4462T:p.R1488W C10orf81 chr C A exonic NM_ :c.C202A:p.L68I TRUB1 chr C T exonic NM_139169:c.C53T:p.T18I FAM204A chr A C exonic NM_ :c.T29G:p.L10R PRLHR chr C T exonic NM_004248:c.G334A:p.A112T C10orf46 chr C A exonic NM_153810:c.G329T:p.S110I EIF3A chr C A exonic NM_003750:c.G2939T:p.R980L EIF3A chr C T exonic NM_003750:c.G1916A:p.R639H SFXN4 chr T C exonic NM_213649:c.A466G:p.S156G BAG3 chr T G exonic NM_004281:c.T624G:p.I208M MCMBP chr G A exonic NM_024834:c.C1073T:p.A358V ATE1 chr T C exonic NM_ :c.A289G:p.M97V TACC2 chr G C exonic NM_006997:c.G1725C:p.Q575H DMBT1 chr C A exonic NM_004406:c.C2983A:p.L995M C10orf120 chr G T exonic NM_ :c.C668A:p.A223D CHST15 chr C G exonic NM_014863:c.G778C:p.G260R CHST15 chr G C exonic stopgain SNV NM_014863:c.C339G:p.Y113X OAT;OAT chr T C NM_ :c.A746G:p.D249G OAT chr T C exonic NM_ :c.A379G:p.I127V ADAM12 chr A G exonic NM_003474:c.T2026C:p.C676R ADAM12 chr G T exonic NM_003474:c.C605A:p.A202E MKI67 chr C T exonic NM_ :c.G4936A:p.V1646I MKI67 chr C G exonic NM_ :c.G3679C:p.D1227H BNIP3 chr A C exonic NM_004052:c.T317G:p.V106G JAKMIP3 chr G C exonic NM_ :c.G1471C:p.E491Q LRRC27 chr T C exonic NM_ :c.T503C:p.L168P KNDC1 chr C T exonic NM_152643:c.C646T:p.R216W VENTX chr C T exonic NM_014468:c.C272T:p.A91V NLRP6 chr C A exonic NM_138329:c.C2530A:p.Q844K IFITM5 chr C T exonic NM_ :c.G268A:p.A90T RNH1 chr C A exonic NM_203384:c.G724T:p.V242L EPS8L2 chr C T exonic NM_022772:c.C323T:p.S108L PNPLA2 chr T G exonic NM_020376:c.T426G:p.N142K AP2A2 chr A G exonic NM_ :c.A99G:p.I33M MUC2 chr C A exonic NM_002457:c.C5249A:p.T1750N 2/11;0/ 19 36/183 ;0/161 51/286 ;4/290 18/81; 0/61 27/110 ;1/110 30/144 ;0/139 46/172 ;0/141 39/234 ;0/171 79/329 ;0/299 24/92; 0/62 45/222 ;0/180 24/109 ;0/89 46/164 ;1/145 21/96; 1/109 22/52; 0/65 30/254 ;1/214 27/90; 0/95 87/336 ;1/385 43/152 ;0/140 34/173 ;0/243 52/66; 0/90 42/54; 1/105 28/115 ;0/106 2/11;0/ 17 18/118 ;2/107 10/72; 1/106 2/12;0/ 25 2/15;0/ 32 68/150 ;0/175 56/103 ;0/107 3/11;0/ 19 23/119 ;2/ /29 4;1/ /91; 0/88 17/85; 1/119 5/22;0/ 31 49/361 ;1/389 10/41; 1/52 23/84; 0/79 89/231 ;0/223 2/18;0/ 19 70/258 ;0/294 42/87; 0/64 52/180 ;0/168 18/63; 0/53 35/122 ;0/117 76/223 ;1/238 60/191 ;0/186 13/63; 0/101 28/115 ;0/116 48/158 ;0/181 3/24;0/ 31 59/175 ;1/137 18/89; 0/84 24/84; 0/84 16/130 ;0/95 58/141 ;0/138 32/105 ;0/92 19/73; 0/87 12/73; 0/81 34/159 ;2/143 19/146 ;0/124 47/241 ;0/265 21/136 ;0/142 30/112 ;1/111 49/201 ;0/246 11/65; 0/97 57/286 ;0/278 2/11;0/ 24 26/157 ;0/143 2/16;0/ 21 8/52;1/ 58 11/81; 0/105 49/353 ;0/392 19/178 ;0/195 15/114 ;0/127 5/44;0/ 48 4/24;0/ 31 78/288 ;0/275 86/272 ;0/313 2/11;0/ 17 17/87; 0/148 20/88; 2/146 35/216 ;0/257 5/36;0/ 34 47/154 ;0/138 4/35;0/ 36 26/115 ;1/107 11/52; 0/36 14/73; 0/80 24/61; 0/53 20/50; 0/42 11/33; 0/36 29/87; 1/118 26/112 ;1/89 26/156 ;0/151 17/130 ;0/121 14/83; 0/99 2/15;0/ 18 17/87; 0/86 40/172 ;1/172 9/39;0/ 46 14/49; 0/88 2/10;0/ 18 61/310 ;0/293 59/157 ;1/182 20/101 ;0/117 20/76; 0/72 11/75; 1/65 17/106 ;0/126 16/73; 0/63 19/95; 0/95 2/16;0/ 24 44/211 ;0/198 39/184 ;0/210 4/38;0/ 35 93/260 ;1/247 75/245 ;2/233 49/207 ;0/226 28/94; 0/131 26/110 ;0/124 19/183 ;1/172 27/111; 0/111 29/165 ;1/163 62/218 ;0/173 24/133 ;0/127 10/22; 0/26 41/123 ;1/147 32/98; 0/90 49/196 ;0/165 12/53; 1/73 19/73; 0/68 11/40; 0/33 17/120 ;0/81 38/195 ;1/203 15/86; 0/112 51/200 ;0/221 15/64; 0/54 28/78; 0/76 7/58;0/ 91 18/148 ;0/253 17/114 ;0/141 18/101 ;0/ /40 6;1/ /62; 0/65 37/157 ;1/173 17/59; 0/49 25/173 ;0/160 33/112 ;1/132 86/282 ;1/250 26/131 ;0/133 16/68; 0/73 10/51; 0/55 18/99; 0/148 13/51; 0/60 12/108 ;1/ /36 9;0/ /127 ;1/124 2/12;0/ 16 24/101 ;0/83 18/80; 0/78 56/230 ;0/206 5/21;0/ 27 11/89; 0/94 24/102 ;0/80 38/176 ;1/214 31/233 ;1/193 15/123 ;0/131 23/90; 0/103 4/19;0/ 22 31/134 ;0/126 2/14;0/ 25 17/67; 0/60 46/169 ;0/147 29/108 ;0/119 55/192 ;0/190
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplemental file 3. All 306 mapped IDs collected by IPA program. Supplemental file 6. The functions and main focused genes in each network.
 LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
TABLE OF CONTENTS Page
 TABLE OF CONTENTS Page Acknowledgements... VI Declaration... VII List of abbreviations... VIII 1. INTRODUCTION... 1 2. BASIC PRINCIPLES... 3 2.1. Hereditary Colorectal Cancer... 3 2.1.1. Differential diagnosis...
TABLE OF CONTENTS Page Acknowledgements... VI Declaration... VII List of abbreviations... VIII 1. INTRODUCTION... 1 2. BASIC PRINCIPLES... 3 2.1. Hereditary Colorectal Cancer... 3 2.1.1. Differential diagnosis...
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Chapter 1. Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes
 Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Best of Uro-oncology Kidney Cancer 2016
 Best of Uro-oncology Kidney Cancer 2016 Ιάσων Κυριαζής, MD, MSc, FEBU Κλινικά εντοπισμένη νόσος 417 PN patients all techniques: Long term oncological outcomes. Only tumor stage was associated with recurrence.
Best of Uro-oncology Kidney Cancer 2016 Ιάσων Κυριαζής, MD, MSc, FEBU Κλινικά εντοπισμένη νόσος 417 PN patients all techniques: Long term oncological outcomes. Only tumor stage was associated with recurrence.
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE ΑΠΟ ΑΤΟΜΑ ΜΕ ΤΥΦΛΩΣΗ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
svari Real-time RT-PCR RSV
 19 39-43 12 Real-time RS svarireal-time RSV subgroup HMPV RSV N Real-time 100 100 Real-time RSV subgroup RSV HMPV F Real-time 55.8 95.5 genotype A2 B1 TaqMan svari InfV RS RSV HMPV HRV PIV RSV HMPV RSV
19 39-43 12 Real-time RS svarireal-time RSV subgroup HMPV RSV N Real-time 100 100 Real-time RSV subgroup RSV HMPV F Real-time 55.8 95.5 genotype A2 B1 TaqMan svari InfV RS RSV HMPV HRV PIV RSV HMPV RSV
J. Dairy Sci. 93: doi: /jds American Dairy Science Association, 2010.
 Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
Si + Al Mg Fe + Mn +Ni Ca rim Ca p.f.u
 .6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
.6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
Identification of Fish Species using DNA Method
 5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
SUPPLEMENTARY INFORMATION
 Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Nature Medicine doi: /nm.2772
 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Supplementary Materials for Evolutionary Multiobjective Optimization Based Multimodal Optimization: Fitness Landscape Approximation and Peak Detection
 IEEE TRANSACTIONS ON EVOLUTIONARY COMPUTATION, VOL. XX, NO. X, XXXX XXXX Supplementary Materials for Evolutionary Multiobjective Optimization Based Multimodal Optimization: Fitness Landscape Approximation
IEEE TRANSACTIONS ON EVOLUTIONARY COMPUTATION, VOL. XX, NO. X, XXXX XXXX Supplementary Materials for Evolutionary Multiobjective Optimization Based Multimodal Optimization: Fitness Landscape Approximation
«Συντήρηση αχλαδιών σε νερό. υπό την παρουσία σπόρων σιναπιού (Sinapis arvensis).»
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
SEN TRONIC AG 3-2 7 0 0 7 A 3 57 3 3 AB 93 :, C,! D 0 7 % 0 7 3 3 93 : 3 A 5 93 :
 # 3-270 07A35733 AB93:,C,!D 07% 0733 93: 3A593:!"#$%% &%&''()*%'+,-. &%&''(/*%'+0. 1*23 '4# 54/%6%7%53 *323 %7 77# %%3#% 8908/"/*55 :1$;/ = 7?@ > 7= 7 %! "$!"#$%&#%'(%%)*#$%&#%'(%#++#,-."/-0-1222"/-0-1
# 3-270 07A35733 AB93:,C,!D 07% 0733 93: 3A593:!"#$%% &%&''()*%'+,-. &%&''(/*%'+0. 1*23 '4# 54/%6%7%53 *323 %7 77# %%3#% 8908/"/*55 :1$;/ = 7?@ > 7= 7 %! "$!"#$%&#%'(%%)*#$%&#%'(%#++#,-."/-0-1222"/-0-1
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Ανάκτηση Εικόνας βάσει Υφής με χρήση Eye Tracker
 Ειδική Ερευνητική Εργασία Ανάκτηση Εικόνας βάσει Υφής με χρήση Eye Tracker ΚΑΡΑΔΗΜΑΣ ΗΛΙΑΣ Α.Μ. 323 Επιβλέπων: Σ. Φωτόπουλος Καθηγητής, Μεταπτυχιακό Πρόγραμμα «Ηλεκτρονική και Υπολογιστές», Τμήμα Φυσικής,
Ειδική Ερευνητική Εργασία Ανάκτηση Εικόνας βάσει Υφής με χρήση Eye Tracker ΚΑΡΑΔΗΜΑΣ ΗΛΙΑΣ Α.Μ. 323 Επιβλέπων: Σ. Φωτόπουλος Καθηγητής, Μεταπτυχιακό Πρόγραμμα «Ηλεκτρονική και Υπολογιστές», Τμήμα Φυσικής,
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supporting Information. Experimental section
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Reminders: linear functions
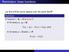 Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
SUPPORTING INFORMATION
 SUPPRTING INFRMATIN Per(-guanidino--deoxy)cyclodextrins: Synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Nikolaos Mourtzis, a Kyriaki Eliadou, a Chrysie Aggelidou,
SUPPRTING INFRMATIN Per(-guanidino--deoxy)cyclodextrins: Synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Nikolaos Mourtzis, a Kyriaki Eliadou, a Chrysie Aggelidou,
Regioselectivity in the Stille coupling reactions of 3,5- dibromo-2-pyrone.
 Regioselectivity in the Stille coupling reactions of 3,5- dibromo-2-pyrone. Won-Suk Kim, Hyung-Jin Kim and Cheon-Gyu Cho Department of Chemistry, Hanyang University, Seoul 133-791, Korea Experimental Section
Regioselectivity in the Stille coupling reactions of 3,5- dibromo-2-pyrone. Won-Suk Kim, Hyung-Jin Kim and Cheon-Gyu Cho Department of Chemistry, Hanyang University, Seoul 133-791, Korea Experimental Section
Mandelamide-Zinc Catalyzed Alkyne Addition to Heteroaromatic Aldehydes
 1 Mandelamide-Zinc Catalyzed Alkyne Addition to Heteroaromatic Aldehydes Gonzalo Blay, Isabel Fernández, Alícia Marco-Aleixandre, and José R. Pedro Departament de Química Orgànica, Facultat de Química,
1 Mandelamide-Zinc Catalyzed Alkyne Addition to Heteroaromatic Aldehydes Gonzalo Blay, Isabel Fernández, Alícia Marco-Aleixandre, and José R. Pedro Departament de Química Orgànica, Facultat de Química,
Supporting Information
 Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)]
![Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)] Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)]](/thumbs/89/98928029.jpg) d Suva refrigerants Technical Information T-9100SI Thermodynamic Properties of Suva 9100 Refrigerant [R-410A (50/50)] Thermodynamic Properties of Suva 9100 Refrigerant SI Units New tables of the thermodynamic
d Suva refrigerants Technical Information T-9100SI Thermodynamic Properties of Suva 9100 Refrigerant [R-410A (50/50)] Thermodynamic Properties of Suva 9100 Refrigerant SI Units New tables of the thermodynamic
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
DuPont Suva. DuPont. Thermodynamic Properties of. Refrigerant (R-410A) Technical Information. refrigerants T-410A ENG
 Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
Single-site association results for 136 SCARB1 genotyped variants with HDL-C.
 Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΕΠΙΔΡΑΣΗ ΥΠΕΡΗΧΩΝ ΣΤΗΝ LISTERIA GRAYI ΣΤΟ ΓΑΛΑ: ΕΠΙΒΙΩΣΗ ΚΑΙ ΦΥΣΙΚΟΧΗΜΙΚΕΣ ΕΠΙΔΡΑΣΕΙΣ. Άρτεμις
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΕΠΙΔΡΑΣΗ ΥΠΕΡΗΧΩΝ ΣΤΗΝ LISTERIA GRAYI ΣΤΟ ΓΑΛΑ: ΕΠΙΒΙΩΣΗ ΚΑΙ ΦΥΣΙΚΟΧΗΜΙΚΕΣ ΕΠΙΔΡΑΣΕΙΣ. Άρτεμις
Malgorzata Korycka-Machala, Marcin Nowosielski, Aneta Kuron, Sebastian Rykowski, Agnieszka Olejniczak, Marcin Hoffmann and Jaroslaw Dziadek
 Molecules 2017, 21, 154; doi:10.3390/molecules22010154 Supplementary Materials: Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effect against
Molecules 2017, 21, 154; doi:10.3390/molecules22010154 Supplementary Materials: Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effect against
Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks
 98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής
 Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Πρόβλημα 1: Αναζήτηση Ελάχιστης/Μέγιστης Τιμής Να γραφεί πρόγραμμα το οποίο δέχεται ως είσοδο μια ακολουθία S από n (n 40) ακέραιους αριθμούς και επιστρέφει ως έξοδο δύο ακολουθίες από θετικούς ακέραιους
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Sampling Basics (1B) Young Won Lim 9/21/13
 Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Solution Series 9. i=1 x i and i=1 x i.
 Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Supplementary Materials for. Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides
 Supplementary Materials for Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides Indrek Kalvet, a Karl J. Bonney, a and Franziska Schoenebeck a * a Institute of Organic
Supplementary Materials for Kinetic and Computational Studies on Pd(I) Dimer- Mediated Halogen Exchange of Aryl Iodides Indrek Kalvet, a Karl J. Bonney, a and Franziska Schoenebeck a * a Institute of Organic
Προσομοίωση BP με το Bizagi Modeler
 Προσομοίωση BP με το Bizagi Modeler Α. Τσαλγατίδου - Γ.-Δ. Κάπος Πρόγραμμα Μεταπτυχιακών Σπουδών Τεχνολογία Διοίκησης Επιχειρησιακών Διαδικασιών 2017-2018 BPMN Simulation with Bizagi Modeler: 4 Levels
Προσομοίωση BP με το Bizagi Modeler Α. Τσαλγατίδου - Γ.-Δ. Κάπος Πρόγραμμα Μεταπτυχιακών Σπουδών Τεχνολογία Διοίκησης Επιχειρησιακών Διαδικασιών 2017-2018 BPMN Simulation with Bizagi Modeler: 4 Levels
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
DuPont Suva 95 Refrigerant
 Technical Information T-95 SI DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 SI DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Supplementary Figure 1. Alternative splicing and cirexon detection in CIRI-AS. (A) Split-alignment of BSJ read pairs in detecting exon skipping,
 Supplementary Figure 1. Alternative splicing and cirexon detection in CIRI-AS. (A) Split-alignment of BSJ read pairs in detecting exon skipping, alternative 5 /3 splicing site and intron retention events.
Supplementary Figure 1. Alternative splicing and cirexon detection in CIRI-AS. (A) Split-alignment of BSJ read pairs in detecting exon skipping, alternative 5 /3 splicing site and intron retention events.
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ. Σπίγγος Γεώργιος
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
Electronic Supplementary Information
 Electronic Supplementary Information NbCl 3 -catalyzed [2+2+2] intermolecular cycloaddition of alkynes and alkenes to 1,3-cyclohexadiene derivatives Yasushi Obora,* Keisuke Takeshita and Yasutaka Ishii*
Electronic Supplementary Information NbCl 3 -catalyzed [2+2+2] intermolecular cycloaddition of alkynes and alkenes to 1,3-cyclohexadiene derivatives Yasushi Obora,* Keisuke Takeshita and Yasutaka Ishii*
DuPont Suva 95 Refrigerant
 Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Th, Ra, Rn, Po, Pb, Bi, & Tl K x-rays. Rn Kα1. Rn Kα2. 93( 227 Th)/Rn Kβ3. Ra Kα2. Po Kα2 /Bi K α1 79( 227 Th)/Po Kα1. Ra Kα1 /Bi K β1.
 Page -1-10 8 10 7 10 6 10 5 10 4 334 ( Th) Counts/Channel 10 3 10 2 10 1 49 ( Th)/ 50 ( Th)/ 50 ( Fr) 0 100 200 300 400 500 600 700 800 900 1000 1000 1100 1200 1300 1400 1500 1600 1700 1800 1900 2000 Channel
Page -1-10 8 10 7 10 6 10 5 10 4 334 ( Th) Counts/Channel 10 3 10 2 10 1 49 ( Th)/ 50 ( Th)/ 50 ( Fr) 0 100 200 300 400 500 600 700 800 900 1000 1000 1100 1200 1300 1400 1500 1600 1700 1800 1900 2000 Channel
Supporting Information
 Supporting Information Montmorillonite KSF-Catalyzed One-pot, Three-component, Aza-Diels- Alder Reactions of Methylenecyclopropanes With Arylaldehydes and Aromatic Amines Li-Xiong Shao and Min Shi* General
Supporting Information Montmorillonite KSF-Catalyzed One-pot, Three-component, Aza-Diels- Alder Reactions of Methylenecyclopropanes With Arylaldehydes and Aromatic Amines Li-Xiong Shao and Min Shi* General
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) CPh 3 as a functional group in P-heterocyclic
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) CPh 3 as a functional group in P-heterocyclic
Conductivity Logging for Thermal Spring Well
 /.,**. 25 +,1- **-- 0/2,,,1- **-- 0/2, +,, +/., +0 /,* Conductivity Logging for Thermal Spring Well Koji SATO +, Tadashi TAKAYA,, Tadashi CHIBA, + Nihon Chika Kenkyuusho Co. Ltd., 0/2,, Hongo, Funabashi,
/.,**. 25 +,1- **-- 0/2,,,1- **-- 0/2, +,, +/., +0 /,* Conductivity Logging for Thermal Spring Well Koji SATO +, Tadashi TAKAYA,, Tadashi CHIBA, + Nihon Chika Kenkyuusho Co. Ltd., 0/2,, Hongo, Funabashi,
Highly enantioselective cascade synthesis of spiropyrazolones. Supporting Information. NMR spectra and HPLC traces
 Highly enantioselective cascade synthesis of spiropyrazolones Alex Zea a, Andrea-Nekane R. Alba a, Andrea Mazzanti b, Albert Moyano a and Ramon Rios a,c * Supporting Information NMR spectra and HPLC traces
Highly enantioselective cascade synthesis of spiropyrazolones Alex Zea a, Andrea-Nekane R. Alba a, Andrea Mazzanti b, Albert Moyano a and Ramon Rios a,c * Supporting Information NMR spectra and HPLC traces
(1) A lecturer at the University College of Applied Sciences in Gaza. Gaza, Palestine, P.O. Box (1514).
 1439 2017, 3,29, 1658 7677: 1439 2017,323 299, 3,29, 1 1438 09 06 ; 1438 02 23 : 33,,,,,,,,,,, : The attitudes of lecturers at the University College of Applied Sciences in Gaza (BA - Diploma) towards
1439 2017, 3,29, 1658 7677: 1439 2017,323 299, 3,29, 1 1438 09 06 ; 1438 02 23 : 33,,,,,,,,,,, : The attitudes of lecturers at the University College of Applied Sciences in Gaza (BA - Diploma) towards
ΑΞΙΟΛΟΓΗΣΗ ΑΦΗΓΗΜΑΤΙΚΩΝ ΙΚΑΝΟΤΗΤΩΝ ΜΕΣΩ ΧΟΡΗΓΗΣΗΣ ΤΟΥ ΕΡΓΑΛΕΙΟΥ ΜΑΙΝ ΣΕ ΤΥΠΙΚΩΣ ΑΝΑΠΤΥΣΣΟΜΕΝΑ ΠΑΙΔΙΑ ΣΤΗΝ ΚΥΠΡΟ
 Σχολή Επιστημών Υγείας Πτυχιακή εργασία ΑΞΙΟΛΟΓΗΣΗ ΑΦΗΓΗΜΑΤΙΚΩΝ ΙΚΑΝΟΤΗΤΩΝ ΜΕΣΩ ΧΟΡΗΓΗΣΗΣ ΤΟΥ ΕΡΓΑΛΕΙΟΥ ΜΑΙΝ ΣΕ ΤΥΠΙΚΩΣ ΑΝΑΠΤΥΣΣΟΜΕΝΑ ΠΑΙΔΙΑ ΣΤΗΝ ΚΥΠΡΟ Γεωργίου Μύρια Λεμεσός, Μάιος 2018 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ
Σχολή Επιστημών Υγείας Πτυχιακή εργασία ΑΞΙΟΛΟΓΗΣΗ ΑΦΗΓΗΜΑΤΙΚΩΝ ΙΚΑΝΟΤΗΤΩΝ ΜΕΣΩ ΧΟΡΗΓΗΣΗΣ ΤΟΥ ΕΡΓΑΛΕΙΟΥ ΜΑΙΝ ΣΕ ΤΥΠΙΚΩΣ ΑΝΑΠΤΥΣΣΟΜΕΝΑ ΠΑΙΔΙΑ ΣΤΗΝ ΚΥΠΡΟ Γεωργίου Μύρια Λεμεσός, Μάιος 2018 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic acids with the assistance of isocyanide
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
Supplementary data. Exome sequencing of hepatoblastoma reveals novel mutations and. cancer genes in the Wnt pathway and ubiquitin ligase complex
 Supplementary data Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex Deshui Jia 1,*, Rui Dong 2,*, Ying Jing 1, 3,*, Dan Xu 1,*,
Supplementary data Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex Deshui Jia 1,*, Rui Dong 2,*, Ying Jing 1, 3,*, Dan Xu 1,*,
Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions
 This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
3.4 SUM AND DIFFERENCE FORMULAS. NOTE: cos(α+β) cos α + cos β cos(α-β) cos α -cos β
 3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Supporting Information. Palladium Complexes with Bulky Diphosphine. Synthesis of (Bio-) Adipic Acid from Pentenoic. Acid Mixtures.
 Supporting Information Palladium Complexes with Bulky Diphosphine Ligands as Highly Selective Catalysts for the Synthesis of (Bio-) Adipic Acid from Pentenoic Acid Mixtures. Choon Heng Low, James D. Nobbs,*
Supporting Information Palladium Complexes with Bulky Diphosphine Ligands as Highly Selective Catalysts for the Synthesis of (Bio-) Adipic Acid from Pentenoic Acid Mixtures. Choon Heng Low, James D. Nobbs,*
ΠΕΡΙΕΧΟΜΕΝΑ. Κεφάλαιο 1: Κεφάλαιο 2: Κεφάλαιο 3:
 4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
E62-TAB AC Series Features
 NEW! E62-TAB AC Series Features Perfect for non-sinusoidal voltages and pulsed s. Housed in a hermetically sealed aluminum can which is filled with environmentally friendly plant oil as standard. The integrated
NEW! E62-TAB AC Series Features Perfect for non-sinusoidal voltages and pulsed s. Housed in a hermetically sealed aluminum can which is filled with environmentally friendly plant oil as standard. The integrated
Supporting Information
 Supporting Information Lewis acid catalyzed ring-opening reactions of methylenecyclopropanes with diphenylphosphine oxide in the presence of sulfur or selenium Min Shi,* Min Jiang and Le-Ping Liu State
Supporting Information Lewis acid catalyzed ring-opening reactions of methylenecyclopropanes with diphenylphosphine oxide in the presence of sulfur or selenium Min Shi,* Min Jiang and Le-Ping Liu State
Lewis Acid Catalyzed Propargylation of Arenes with O-Propargyl Trichloroacetimidate: Synthesis of 1,3-Diarylpropynes
 Supporting Information for Lewis Acid Catalyzed Propargylation of Arenes with O-Propargyl Trichloroacetimidate: Synthesis of 1,3-Diarylpropynes Changkun Li and Jianbo Wang* Beijing National Laboratory
Supporting Information for Lewis Acid Catalyzed Propargylation of Arenes with O-Propargyl Trichloroacetimidate: Synthesis of 1,3-Diarylpropynes Changkun Li and Jianbo Wang* Beijing National Laboratory
Μηχανική Μάθηση Hypothesis Testing
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Μηχανική Μάθηση Hypothesis Testing Γιώργος Μπορμπουδάκης Τμήμα Επιστήμης Υπολογιστών Procedure 1. Form the null (H 0 ) and alternative (H 1 ) hypothesis 2. Consider
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Μηχανική Μάθηση Hypothesis Testing Γιώργος Μπορμπουδάκης Τμήμα Επιστήμης Υπολογιστών Procedure 1. Form the null (H 0 ) and alternative (H 1 ) hypothesis 2. Consider
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
Supplementary information:
 Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Homework 3 Solutions
 Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
DETERMINATION OF THERMAL PERFORMANCE OF GLAZED LIQUID HEATING SOLAR COLLECTORS
 ΕΘΝΙΚΟ ΚΕΝΤΡΟ ΕΡΕΥΝΑΣ ΦΥΣΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΗΜΟΚΡΙΤΟΣ / DEMOKRITOS NATIONAL CENTER FOR SCIENTIFIC RESEARCH ΕΡΓΑΣΤΗΡΙΟ ΟΚΙΜΩΝ ΗΛΙΑΚΩΝ & ΑΛΛΩΝ ΕΝΕΡΓΕΙΑΚΩΝ ΣΥΣΤΗΜΑΤΩΝ LABORATORY OF TESTIN SOLAR & OTHER ENERY
ΕΘΝΙΚΟ ΚΕΝΤΡΟ ΕΡΕΥΝΑΣ ΦΥΣΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΗΜΟΚΡΙΤΟΣ / DEMOKRITOS NATIONAL CENTER FOR SCIENTIFIC RESEARCH ΕΡΓΑΣΤΗΡΙΟ ΟΚΙΜΩΝ ΗΛΙΑΚΩΝ & ΑΛΛΩΝ ΕΝΕΡΓΕΙΑΚΩΝ ΣΥΣΤΗΜΑΤΩΝ LABORATORY OF TESTIN SOLAR & OTHER ENERY
Συστήματα Διαχείρισης Βάσεων Δεδομένων
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Συστήματα Διαχείρισης Βάσεων Δεδομένων Φροντιστήριο 9: Transactions - part 1 Δημήτρης Πλεξουσάκης Τμήμα Επιστήμης Υπολογιστών Tutorial on Undo, Redo and Undo/Redo
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Συστήματα Διαχείρισης Βάσεων Δεδομένων Φροντιστήριο 9: Transactions - part 1 Δημήτρης Πλεξουσάκης Τμήμα Επιστήμης Υπολογιστών Tutorial on Undo, Redo and Undo/Redo
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
