Nature Medicine doi: /nm.2772
|
|
|
- Κυρία Αλαφούζος
- 7 χρόνια πριν
- Προβολές:
Transcript
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value* vs. 4OHT Target SLC2A ANKRD E E E FSIP2 n.s. n.s SLC1A E 04 n.s E C10orf n.s MCOLN IQGAP E E TMPRSS11F n.s LCA CPA4 n.s CYP24A n.s KTN RB1CC CD2AP LOC RIMKLB FRMD6 n.s KRT n.s JMJD1C n.s NIPBL HSPA4L E CLEC2B n.s DAB EMP (b) NAV ARHGAP E E BMPR1B CCDC KLHL n.s GPR RARB n.s INPP4B RECQL HIST1H2AL n.s PIK3CA n.s E (b) GBP3 n.s MPP5 n.s GOLGB n.s NCRNA n.s ACTA PNPLA n.s NSAP E 06 n.s E PRDM E KLHL KRTAP NT5E n.s E C13orf LPHN E 06 n.s E WWC CTNNAL FRYL n.s GOPC n.s E ATP13A PLK E CLDN E DCP HNMT n.s E RICTOR n.s ARHGAP n.s
23 STAMBPL CFTR FCHO n.s HIPK E KRTAP SGTB n.s L3MBTL E E SLC7A E E CLDN n.s DAAM n.s RASA n.s VSNL n.s CTGF E E RAB3B n.s E NEIL n.s HBEGF n.s E MBNL n.s TMEM ALS2CR n.s YOD WDR n.s XRN n.s TFPI E E PFDN n.s TBC1D PDLIM STARD n.s SPTLC E CRIM n.s RAB KRTAP TEAD GCNT E E UTRN E HAPLN n.s PCMTD HEG E POC1B FMNL IMPA FBXO n.s HEATR5B n.s ITGA ARHGAP NFE2L n.s NDFIP n.s CAV OSGIN E CDC14A E E (b) STK17A CRIPT n.s FEM1C E SEPP E E TSPAN n.s PLEKHA LGR n.s (a) E2F n.s KRTAP3 1 n.s E RP n.s IL n.s SNORD73A n.s CD n.s GDA n.s E
24 CHIC n.s SYNE n.s ZNF n.s SLC28A E 04 n.s E UBE2D E 04 n.s E CAV E E (b) RALGPS E 04 n.s E CYP39A n.s YPEL ABCA E 09 n.s E CRY TJP JPH n.s PTPN ERV n.s DOCK TOR1AIP LGALS n.s E CHMP4C n.s CCDC RBMS n.s RIOK E E RAET1L n.s SERPINB n.s (b) TIMP (b) TES E E RNF FAM134B E 06 n.s E UBN E E TMX n.s SESN E E CLK USP RGL n.s PMAIP E E WHAMM n.s PLCE n.s ODZ n.s CYBRD E E SAV n.s GFPT DOCK SNAP n.s HIST1H2BM n.s XK ZFAND n.s ITGB n.s ARNTL PPFIBP NETO n.s TTPA DUSP (b) BRD n.s GPR137B n.s GAB TLR n.s BAG (a) SWAP n.s PARD6B n.s E DOCK n.s E UBL n.s PAWR TGFBR
25 ERO1LB n.s TGFA n.s (b) SLC1A E E SOS n.s PIM n.s KLF n.s TNFRSF n.s (b) AFF E 04 n.s E AVIL n.s SNORD BMP ANK n.s RRS n.s TM4SF E TRIP n.s POLR1B E E (a) THBS SEMA3C n.s (b) RTN n.s CLTB ZFAND IL22RA n.s C1orf n.s KLF E 04 n.s E (b) HSD17B E E NEDD4L n.s (b) CDKN1B n.s (c) ARID5B (a) RNF MAGI n.s SLC31A E 05 n.s E (b) GADD45B TRAK n.s CORO1A (b) KLF n.s GPR TGFBR n.s ABLIM n.s JAK CYR E E (b) CCNG (b)(c) C18orf n.s KLK (b) CDKN2B n.s FOXP E E LIPG GATS SNORD MTNR1A CDCA (a) LDLR E 04 n.s E ABHD n.s SAT TSC n.s ERRFI n.s NPIPL n.s NCS ATXN E E TCF n.s ATL E E FAM27E n.s KIT SRGAP n.s
26 CENPP TNS n.s GLUL n.s SH3TC n.s SMPDL3A (b) ZRANB CITED E E HECA n.s STX UBE2H (b) ATP8A E 04 n.s E EGR E E SNX n.s EDN (b) PIGM n.s PDIA DDI n.s EIF2C n.s ARHGEF E 04 n.s E NT5C n.s DPYSL IL1F E E LIPH FMO TMEM E 04 n.s E CYB5RL n.s TSR SLC12A E 04 n.s E DUSP E E (b) GEMIN n.s (a) BRAP n.s TMEM200A n.s WIPI RRP1B n.s FCHSD n.s C10orf n.s PTPRJ E 05 n.s E NOP n.s BCAS WDFY n.s ACPP TMEM n.s EFNA n.s NPC n.s RRP n.s NDP MACF n.s TDRD OPTN RASGRP CDCA7L FKBP POLR1A n.s BYSL n.s (a) CTH USP ARL2BP SLC5A n.s TMEM86A MXI E 04 n.s E (b) SASH n.s SCIN E 07 n.s E RAB5B
27 NPNT n.s FAM59A n.s LRP E E CTSB E (b) S100A n.s (b) OR7E87P PSMG n.s GRIK NTAN E 04 n.s E SQLE TIMM8B n.s E SORL E E PPP1R15A MRPL n.s B4GALNT n.s FOXO n.s MAML PYCR n.s C10orf n.s MRPL n.s DSP NME n.s UCA RAB9A n.s NRM GPLD n.s PELI MAOB E E SLC4A FOSL E 04 n.s E TBC1D E E CHMP4B E 05 n.s E GATSL KIF13B n.s CTDSP ATP9A NECAP n.s RERE E (b) LGR E 04 n.s E EGLN n.s SUN n.s ARRDC E E TRIM n.s AIMP E FGF E 04 n.s E ZFHX LY6G6C n.s PRR MFSD SLC29A n.s C8orf n.s MTSS1L TGFB PAQR n.s S100P n.s (b) IRS E 06 n.s E EPHX n.s REXO E 05 n.s E MAST DNMBP SCD n.s (a) SLC2A n.s CCDC
28 BTG n.s (c) ANP32A n.s PLXNB n.s MBTPS SPINK E PDK MEGF E 04 n.s TST (b) HBP BCL E E (b)(c) AKAP n.s BTG n.s NFIC n.s NDRG E E IPO n.s BACH E 05 n.s E IGF1R n.s (b) SLC16A n.s KIF3C (b) C17orf n.s SLC6A TMEM161A TNFSF E E (c) GYLTL1B n.s BOD n.s KPNA E 05 n.s E SNORD CARS n.s TMEM E EPB41L n.s TTLL n.s FOXO TEP n.s CYTH C19orf n.s ZFYVE E 04 n.s GMPR GAB E CHD n.s SLC19A n.s (a) NDUFV ERBB E E (b) RNF LNX E IGFL E WDR n.s (a) TNRC SLC16A GTPBP n.s TUBA1A n.s IER E 05 n.s CBLB TMEM n.s AMPD (a) GRB E (b) HES SLC2A n.s RNF n.s TMEM E 06 n.s E SLC44A ATG2A TIGD n.s TRERF
29 SLC45A n.s SOBP n.s PPAP2B E E RNF IL10RA n.s CCL PNPLA INSIG E ARHGAP n.s GTF2I n.s (b) VAMP LAD n.s PIK3IP E 04 n.s SH3RF n.s INSR E 04 n.s ZNRF n.s (a) RAB MPI TCP11L E E CARHSP E MID1IP E YPEL CABLES n.s EPN ULK SLC43A E ZNF n.s GSN (b) PPFIBP TNS n.s MED n.s FASN CALCOCO LAMC REPS SCNN1B n.s TMCC REG n.s THOC CA5B n.s TLCD DBP E PLCG E 05 n.s E (b) PIK3C2B C10orf E 05 n.s E SGSH NDUFB FBXO E 04 n.s E (c) PDZK1IP TP53INP SULT2B ID (a) STARD ZNF n.s SLC1A (b) LFNG n.s TNFRSF EFNA DGCR ZNF n.s DAPK E (b) TNFSF SERPIND
30 GPR SOD (c) SLC22A n.s HSD17B n.s PRR15L ASCL E (a) KRTAP CDX SULT1C E TXNIP TRIM RND E DHRS TMOD n.s PLA2G2F n.s Supplementary Table 1. High Stringency Gene List of Genes Regulated at 12 h Treatment. The results of the microarray analysis were cut off at a significance level of Bonferroni adjusted P value (asterisk) 0.05 (corresponding to a false discovery rate (FDR) P value ) and a fold change (FC) of at least two fold in at least one of the treatments. The fold change is displayed as value in treated cells versus value in control cells (vehicle). n.s. = not significant in a two tailed ANOVA analysis (P value > 0.05). The list is sorted indicating first the highest fold change value for double treatment (4OHT+DOX) versus 4OHT. Known target genes were previously described: (a) gene found to be regulated by N TCF (van der Flier et al., 2007); (b) FoxO3a target gene described by Delpuech et al., 2007 and (c) summarized by Greer and Brunet, 2005.
31 Supplementary Table 2: FOXO3a and/or beta catenin regulated genes (24 h treatment) 4OHT DOX 4OHT+DOX FC 4OHT+DOX Known Gene Symbol FC P value* FC P value* FC P value* vs. 4OHT Target SLC2A E SLC1A n.s KRTAP n.s CPA E E E ANKRD E E E DAB E IQGAP E E RTKN KRTAP n.s CLDN n.s WWC DEPDC n.s KIF18A n.s DIAPH PRDM E SEMA5A LPHN n.s MS4A n.s SPINK E ULBP n.s CLDN n.s ARHGAP11A n.s KRTAP C13orf CLEC2D n.s SLC31A (b) STARD PHLDB BRCA n.s SLC28A n.s E ATAD n.s NSAP n.s SLC45A E 04 n.s E SDR16C n.s EMP E (b) PSG n.s JMJD1C n.s HEG E CP n.s SELP E 04 n.s E AMOTL E EDN E E (b) PUS7L n.s ZBED n.s AXL n.s NAV REEP n.s VSIG PIK3CA n.s E (b) C4orf n.s RIMKLB SKIL n.s NPIPL n.s E DUT MAGI n.s TNC GCNT E RAB3B n.s ANLN
32 HAUS n.s MIR RAD51AP n.s CTGF E E COL10A n.s DSCC ABCA E 05 n.s E STX RARB n.s FCER1A n.s NBEAL PSG n.s XG n.s WDHD E 04 n.s SASH n.s PLK E MMP n.s RICTOR n.s CCDC n.s IL1F E E GADD45B NTAN n.s MALT KIF LAMA n.s DEPDC1B n.s GPRC5A MAP E TINAGL E CLIC n.s TBC1D YOD KPNA E E C21orf n.s E CTNNAL RAB n.s SEMA3A n.s TIMELESS n.s GREB n.s E FGD n.s ARHGAP FBXO (c) WDR n.s ZDHHC NTN n.s C6orf n.s SCML ZSWIM n.s TNFAIP TFPI n.s CSRNP LY6G6E n.s MPHOSPH n.s MBNL n.s FOS CCDC CHN n.s HELLS CDKN2B EIF1AY n.s MAML FCHSD n.s TUFT n.s
33 TBC1D n.s E TMEM E E CGA n.s TEK n.s SLC7A E E RAD n.s CSGALNACT n.s SYNE NEBL n.s PTPRR n.s FYN E GTSE n.s TMSB4X n.s CYR E E (b) FRYL n.s RGL E AMIGO SLC1A TTC n.s NT5E n.s DOCK SERPINB (b) PYGL CRIM E SECISBP2L n.s FBXO n.s E FANCA n.s XRN n.s RALGPS E E ALS2CR PLOD E E CEP NOC3L E (a) TEAD BLM n.s MCTP E E C5orf E BACH OSGIN n.s DAAM n.s PRC n.s (b) CLTB ELOVL n.s CEP n.s GULP n.s VILL n.s SLC30A n.s DUSP n.s HBEGF n.s CYTL n.s UBE2B n.s SYNJ n.s RAD L3MBTL n.s E ABTB n.s E TIMP n.s (b) PMAIP n.s PHACTR n.s BAMBI n.s ATL PBK n.s RAB5A n.s PEX n.s
34 CLDN FCHO HIST1H1B n.s ZNF E 04 n.s E ITGB n.s WDFY E FABP SCIN E 04 n.s E MARCH n.s HECA n.s GTF2B n.s PIK3AP CTSL POC1B TMSL n.s ATP6V1E n.s CYB5R KIF n.s (b) CYTH n.s AURKA n.s (b) RAB4B n.s E INPP4B n.s DDX CAV E E (b) POLH n.s MFGE E 04 n.s E RASA n.s TRAM n.s SLC7A E (b) NUSAP n.s IL n.s E CA5B E XPO n.s PLEKHA n.s MMD HIST2H3A n.s DNAJB PPP2CB CAV HIST1H4K E PHGDH C11orf n.s APLF n.s PARD6B n.s SESTD n.s TIMP n.s GSG BUB E 04 n.s CHEK NKD SLC7A E (a) HAPLN n.s FANK n.s EZH MSH SYTL n.s RNF n.s CHMP4C n.s SLC2A MXI n.s (b) PAWR n.s TCF AURKB (a)
35 SKA n.s PIK3IP n.s SHCBP n.s OBFC2A n.s ODZ E E RFC n.s SNHG n.s STXBP UBE2C (b) EIF2C UPK3B SUV39H n.s WDR n.s (b) PDLIM MRPS n.s SESN ATG2A NEK n.s (b) BACH E 04 n.s E TSPAN UBE2D n.s CDC14A E E (b) INTS n.s CDKN1A n.s (c) PCMTD MXRA n.s SCARA SSFA n.s TRAK CHRNB RAD54L n.s RIOK n.s RAD51C n.s SNORD4A n.s UTP RTN n.s TDRD UCHL E RAET1L DDX (a) LMO n.s CYTH n.s UCK n.s NAV GFPT n.s RNF n.s SNAP E 05 n.s E IRF CHIC E 04 n.s E RSPH n.s TES E E PPIH NOL NUDT E E SPAG n.s XPR n.s SWAP n.s SEC14L n.s AFF E 05 n.s E ZYX HIST1H2BM n.s OSGEPL n.s RFC
36 HIVEP E C15orf SP KLF n.s ZFHX TJP n.s TMOD n.s MBNL n.s RANBP B4GALNT CCNG (b)(c) PTPN E E EPS8L n.s TROAP MAP1LC3B E PTPN n.s E CDCA CYP39A E E NDRG E E SMURF n.s OR2A20P E 05 n.s E KDELC n.s AGR E SLC29A n.s FLJ DOCK n.s SNORD ZFYVE E E WDTC ACVR1C n.s NUP n.s CR SEMA3C (b) ZBTB n.s FANCC n.s FOXM (b) NECAP n.s WWTR n.s SLC1A n.s CCDC PTPN CHMP n.s PPFIBP n.s ILDR ETHE ATP8A NCAPD E 04 n.s GATA n.s PDGFRL TCP11L TMEM E 06 n.s E TNS n.s SMC1A E SHROOM E E RAB9A n.s CD E 04 n.s E IL2RG E 04 n.s E LDLR ARID5B E (a) TCF n.s GATSL E E ITGA n.s CD (b)
37 TMPO n.s RACGAP n.s FANCG MCM E ANXA ABLIM n.s PIK3C n.s SLC12A SUN E 05 n.s E BACE POLB E KLHL SNX n.s NAPG POLD MEGF GBE n.s GM2A (b) SLC4A n.s DCAF CRIPT n.s WDSUB n.s NSUN5P ZFAND n.s HIST1H3I n.s REEP n.s KIF18B E E C9orf n.s ERV HADH n.s QPCTL n.s SKP n.s RRM n.s CBLB CALCOCO SPTB ZFAND E E STX ICK E E RCCD ARHGEF n.s REPS CALHM n.s RBMS MTO SPTAN TP n.s TSR E PC n.s CXCL n.s (a) LASS C1orf KIF3C (b) TMEM86A KLF NDUFV n.s GAB n.s TYMS n.s TNFRSF E E (b) TEP CCDC n.s E2F n.s ANAPC n.s
38 SEPP E E ACPP E OPTN n.s BCL (b)(c) TGFBR n.s FEN n.s RCAN TUBA1A n.s STRADB E E LASS n.s GDA n.s SAT E E TOR1AIP n.s NAF LAMC RYBP n.s YIPF FRMD n.s OTUD n.s PTPRG E E MAGI n.s NT5C n.s JAK E E GMDS n.s NUP n.s LY6G6C E E FOXP E E C7orf n.s CYBRD E 04 n.s E ARHGEF E 04 n.s E IPO n.s PRSS n.s TTF n.s PPAT TRMU n.s XRCC6BP n.s LRRC8A PINX n.s NCAPH ESPL POU5F n.s FMNL n.s NHLRC PNRC YPEL URB n.s UCA n.s DDI KIAA n.s ATXN n.s BTG n.s (c) TIPARP n.s CYTH n.s RAB ANGPT E 04 n.s E BMPR VEPH n.s ARHGAP n.s HSD17B n.s FOXO E E RHOF IGFBP GEMIN (a)
39 ABHD n.s TPRA E 04 n.s E FADS HSPE n.s FLNB n.s E TUBG n.s SLC5A n.s BUB1B n.s VAMP n.s PDSS KLF n.s (b) PRR MACF E 04 n.s E DNMT n.s NOP n.s RNFT n.s RND n.s NEDD4L n.s (b) PIGW n.s (a) DTL n.s GPR137B n.s CDC n.s RRP SERINC n.s PHF21A PIK3CB n.s PDZK1IP PPP1R15A PDIA E E HSPA4L n.s MAP4K n.s CCNDBP GATS LGR n.s SOD n.s (c) TRIM FOSL E 04 n.s E KIF20A E E TSPAN E E FGF E 04 n.s E MCM n.s CITED n.s C10orf n.s LRP SLC2A E 05 n.s E SLC35D ITGAX TCOF ERBB E E (b) ARRDC E E DPYSL E 05 n.s E CENPM E 05 n.s E RERE n.s (b) TGFA n.s (b) FAM64A n.s EPRS n.s LGMN TMCC E SRM (a) ABI n.s GPX n.s LGR n.s (a) DUSP E (b)
40 PLK SFMBT n.s FARSB n.s IARS n.s (b) CD OR2A n.s GALNT n.s TM4SF E E TTC SUPT16H n.s PTTG1IP n.s OR2A n.s PSRC E2F n.s SHMT n.s RBMX n.s ZNF n.s SPAG SNORA CRTAM TGFBR TBC1D n.s XPO TRNP n.s ITGB CAD n.s (a) PAK1IP DUSP E E (b) CTSB E 04 n.s E (b) IRS E 05 n.s E DHRS n.s ERRFI E E EGLN SCFD n.s ACY COL17A C10orf APCDD n.s UCP CYP2S E E PPIF E (a) TNS E E CGREF n.s HLA E n.s UBE2H n.s (b) GPRC6A KCTD E E PPAP2B E SLC2A PSMG n.s BAG n.s (a) SLC43A E 04 n.s E PYCR RPL27A n.s GDF MDC n.s PXK n.s CASZ HSPG NUCKS n.s TMEM NAT n.s GPR
41 KIFC n.s PLAT n.s ATP5G n.s MAST ACACA n.s CAMK2N MMACHC n.s WRB n.s AKAP n.s TOE n.s E USP HNMT C5orf n.s LEF E 04 n.s E (a) NR6A n.s TNRC MRPS n.s SORL TNFSF E E (c) MRGPRX E E TARBP n.s C19orf n.s CARS n.s CLIC TRIP n.s UCK n.s ANGEL n.s NUP n.s PCDH PRPF n.s KLK n.s (b) SREBF STEAP SERPINE E E RRP1B ATP9A E E EPHX S100P E 05 n.s E (b) RAB11FIP GEMIN E E (a) CAB39L TP53INP E E NLE n.s (a) ITLN n.s TOMM40L WDR n.s OAS LIPH IL10RA E 05 n.s E MRPL n.s (a) GRB (b) DKC n.s (a) ITM2B NFX n.s GLUL E E DLEU n.s FSTL E 04 n.s E MAOB E 05 n.s E PRMT n.s THBS E E ILF n.s WDR (a) SLC1A E E (b)
42 NBEAL n.s HES KIF13B n.s AADAT n.s CCDC n.s WDR n.s CLU n.s (b) TNFRSF EGFR n.s SF3B n.s PLBD AFF ABHD ASF1A n.s KSR n.s CDC42EP n.s (b) GPI n.s NRM n.s E DCAKD n.s RAG1AP MAZ ZMYM SLC16A n.s MLEC n.s TUBA4A n.s AFAP n.s E SLCO4A n.s TSPAN E E E KIF2C n.s UTP FRRS n.s MT1X n.s SLC44A MAGT MANEAL n.s E PCYOX1L n.s UTP14A LRRC37A LGALS3BP PA2G E 05 n.s E GNL n.s AIMP n.s CDK n.s (a) MRPL n.s IGF1R E E (b) PLCE n.s E PRKAR2A ARHGAP E 04 n.s GSN (b) CTTNBP n.s CD (b) HGSNAT CLDN (a) HSD17B n.s CRAT E E SCARB n.s TNS COQ MID1IP n.s ATP1B LPHN n.s MOBKL2B n.s APRT
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
Prostate-targeted radiosensitization via aptamer-shrna chimeras in human. tumor xenografts
 SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
500 New CUSABIO Antibodies at MedStore!
 BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in. and *Christina Chan
 Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
Supplementary Online Content
 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent
 Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Burden of Disease Variants in Participants of the Long Life Family Study. Supplemental Online Material
 Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
 Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT
 Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis
 S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
Supplementary Information for. RRM2B Suppresses Activation of the Oxidative Stress Pathway and is. Up-regulated by P53 During Senescence
 Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Materials for
 www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
NCBI ID Symbol 2 A2M A4GALT A4GNT 8086 AAAS 13 AADAC AADACL AAGAB 15 AANAT 16 AARS AARS AASS 18 ABAT 19 ABCA1
 NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
Supplementary Table S1 Demographic characteristics of HCC patients
 Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Materials for
 www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells
 Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Supplementary Figure 1.
 Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal
 Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplementary Materials for
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Supplementary Materials for PD-1 Marks Dysfunctional Regulatory T cells in Malignant Gliomas Daniel E. Lowther 1,, Brittany A. Goods 2,, Liliana E. Lucca 1,, Benjamin A.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 Supplementary Materials for PD-1 Marks Dysfunctional Regulatory T cells in Malignant Gliomas Daniel E. Lowther 1,, Brittany A. Goods 2,, Liliana E. Lucca 1,, Benjamin A.
Zhang et al., Supplemental figures, Methods, References and Tables
 1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
Supplemental Information. A p53 Super-tumor Suppressor Reveals a Tumor. Suppressive p53-ptpn14-yap Axis. in Pancreatic Cancer
 Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in
 1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
Η γενετική πολυπλοκότητα του καρκίνου του παγκρέατος
 Η γενετική πολυπλοκότητα του καρκίνου του παγκρέατος Δρ Αθανάσιος Αθανασιάδης Αθανάσιος Αθανασιάδης Δεν έχω αντικρουόμενα συμφέροντα προς γνωστοποίηση Καρκίνος παγκρέατος Θνητότης Επίπτωση Καθυστερημένη
Η γενετική πολυπλοκότητα του καρκίνου του παγκρέατος Δρ Αθανάσιος Αθανασιάδης Αθανάσιος Αθανασιάδης Δεν έχω αντικρουόμενα συμφέροντα προς γνωστοποίηση Καρκίνος παγκρέατος Θνητότης Επίπτωση Καθυστερημένη
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros
SUPPLEMENTARY INFORMATION doi:10.1038/nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros
Comparative transcriptome analysis of peripheral blood mononuclear cells in
 Comparative transcriptome analysis of peripheral blood mononuclear cells in hepatitis B-related acute-on-chronic liver failure Qian Zhou, Wenchao Ding, Longyan Jiang, Jiaojiao Xin, Tianzhou Wu, Dongyan
Comparative transcriptome analysis of peripheral blood mononuclear cells in hepatitis B-related acute-on-chronic liver failure Qian Zhou, Wenchao Ding, Longyan Jiang, Jiaojiao Xin, Tianzhou Wu, Dongyan
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein
 1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
Analysis Summary: Any Allergy
 Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion
 Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
Supplementary Table 1. Composition of the diets
 Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Information
 Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
GeneTex Sonderaktion: Custom Antibody Panels
 (Stand 01/2016) GeneTex Sonderaktion: Custom Antibody Panels Fünf Antikörper (in der 25 µl-größe) zum Sonderpreis von nur 420,00 (statt 545,00 ). Bitte geben Sie bei Ihrer Bestellung die Angebotsnummer
(Stand 01/2016) GeneTex Sonderaktion: Custom Antibody Panels Fünf Antikörper (in der 25 µl-größe) zum Sonderpreis von nur 420,00 (statt 545,00 ). Bitte geben Sie bei Ihrer Bestellung die Angebotsnummer
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma. Supplement Figure 1. Regulation of PGC1α expression in cultured podocytes.
 Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Supplemental Materials and Results
 Supplementary Figures and Legends Supplemental Materials and Results Supplementary Figure 1. Metabolic parameters in Ad-mKO mice A. TG content in skeletal muscle of Ctrl and Ad-mKO mice on chow diet (CD)
Supplementary Figures and Legends Supplemental Materials and Results Supplementary Figure 1. Metabolic parameters in Ad-mKO mice A. TG content in skeletal muscle of Ctrl and Ad-mKO mice on chow diet (CD)
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Single-site association results for 136 SCARB1 genotyped variants with HDL-C.
 Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
(A) Rounds of replating (R) of MLL-AF9 transformed LSK colonies from initiation
 upplemental Figure. (A) Rounds of replating (R) of LL-AF transformed LK colonies from initiation experiments (.E. of triplicate experiments, p=.)., p
upplemental Figure. (A) Rounds of replating (R) of LL-AF transformed LK colonies from initiation experiments (.E. of triplicate experiments, p=.)., p
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
In utero vitamin D deficiency increases airway smooth muscle mass and impairs lung
 1 Supplemental Information 2 Title 3 4 In utero vitamin D deficiency increases airway smooth muscle mass and impairs lung function 5 Authors 6 7 1 Rachel E Foong, 1 Anthony Bosco, 1 Anya C Jones, 1 Alex
1 Supplemental Information 2 Title 3 4 In utero vitamin D deficiency increases airway smooth muscle mass and impairs lung function 5 Authors 6 7 1 Rachel E Foong, 1 Anthony Bosco, 1 Anya C Jones, 1 Alex
ΤΖΕΤΗ Μ.
 ΠΡΟΓΕΝΝΗΤΙΚΟΣ ΕΛΕΓΧΟΣ Πως η εφαρμογή στα μεταγεννητικά δείγματα διευκόλυνε την ΠΓΔ ΔΟΜΗ Εισαγωγή arrays/ CNVs-[MCA: molecular karyotype analysis] Είδη arrays, διαφορές Ανάλυση/ επεξήγηση αποτελεσμάτων
ΠΡΟΓΕΝΝΗΤΙΚΟΣ ΕΛΕΓΧΟΣ Πως η εφαρμογή στα μεταγεννητικά δείγματα διευκόλυνε την ΠΓΔ ΔΟΜΗ Εισαγωγή arrays/ CNVs-[MCA: molecular karyotype analysis] Είδη arrays, διαφορές Ανάλυση/ επεξήγηση αποτελεσμάτων
Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated
 Supplemental Figure legends Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated genes. a) Mitotic roles of Polo-like kinase. b) Cell cycle regulation by BTG
Supplemental Figure legends Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated genes. a) Mitotic roles of Polo-like kinase. b) Cell cycle regulation by BTG
ΚΛΑΙΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ ΜΟΡΙΑΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ ΜΟΡΙΑΚΗ ΓΕΝΕΣΙΚΗ ΜΟΡΙΑΚΗ ΜΙΚΡΟΒΙΟΛΟΓΙΑ. Χρόνοσ Απάντθςθσ
 ΚΛΑΙΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ Χρωμοςωμικι ανάλυςθ αμνιακοφ υγροφ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ CVS (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ εμβρυϊκοφ αίματοσ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ
ΚΛΑΙΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ Χρωμοςωμικι ανάλυςθ αμνιακοφ υγροφ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ CVS (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ εμβρυϊκοφ αίματοσ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ
Aluminum Electrolytic Capacitors
 Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Supporting Information. Property focused structure-based optimization of small molecule inhibitors of the proteinprotein
 Supporting Information Property focused structure-based optimization of small molecule inhibitors of the proteinprotein interaction between menin and Mixed Lineage Leukemia (MLL) Dmitry Borkin a, Jonathan
Supporting Information Property focused structure-based optimization of small molecule inhibitors of the proteinprotein interaction between menin and Mixed Lineage Leukemia (MLL) Dmitry Borkin a, Jonathan
Aluminum Electrolytic Capacitors (Large Can Type)
 Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Nature Immunology: doi: /ni Supplementary Figure 1. Foxm1 loss leads to bone marrow hypocellularity.
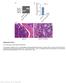 Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
http / / cjbmb. bjmu. edu. cn Chinese Journal of Biochemistry and Molecular Biology RT-PCR Runx d PCR RT-qPCR Western BMPs Wnt Ihh
 ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 6 27 6 540 ~ 547 micrornas * / / 250062 microrna mrna β- RT-PCR Runx2 0 7 14 d micrornas
ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 6 27 6 540 ~ 547 micrornas * / / 250062 microrna mrna β- RT-PCR Runx2 0 7 14 d micrornas
Original Article Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells
 Int J Clin Exp Med 2014;7(11):4253-4259 www.ijcem.com /ISSN:1940-5901/IJCEM0002259 Original Article Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells Chen Chen 1, Yachun
Int J Clin Exp Med 2014;7(11):4253-4259 www.ijcem.com /ISSN:1940-5901/IJCEM0002259 Original Article Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells Chen Chen 1, Yachun
Unique peptides 1 DHX9 ATP-dependent RNA Q MYBBP1A Myb-binding protein 1A Q9BQG ADAR Adenosine deaminase, Q59EC
 SUPPLEMENTAL TABLES Supplemental Table 1: List of proteins identified by mass spectrometry. # Symbol Characteristics of proteins Accession number Molecular weight (kda) Unique peptides 1 DHX9 ATP-dependent
SUPPLEMENTAL TABLES Supplemental Table 1: List of proteins identified by mass spectrometry. # Symbol Characteristics of proteins Accession number Molecular weight (kda) Unique peptides 1 DHX9 ATP-dependent
Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia
 SUPPLEMENTAL MATERIAL FOR: Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia Matthew T. Witkowski, Luisa Cimmino, Yifang Hu, Thomas Trimarchi,
SUPPLEMENTAL MATERIAL FOR: Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia Matthew T. Witkowski, Luisa Cimmino, Yifang Hu, Thomas Trimarchi,
 Supplemental Figure 1 Genechip data analysis. Three pairs of bulge and sub-bulge ORS Genechips were analyzed and compared by MASv5.0 and dchip 1.3 software algorithms in order to identify gene transcripts
Supplemental Figure 1 Genechip data analysis. Three pairs of bulge and sub-bulge ORS Genechips were analyzed and compared by MASv5.0 and dchip 1.3 software algorithms in order to identify gene transcripts
Validità 31/01/2013 PROMO CODE
 Validità 31/01/2013 PROMO CODE SCP-P100192 SC Cat # Protein Name Application Y71-30G 14-3-3α/β Protein Western Blot Y71-30N 14-3-3α/β Protein Western Blot Y75-30G 14-3-3ε Protein Western Blot Y75-30N 14-3-3ε
Validità 31/01/2013 PROMO CODE SCP-P100192 SC Cat # Protein Name Application Y71-30G 14-3-3α/β Protein Western Blot Y71-30N 14-3-3α/β Protein Western Blot Y75-30G 14-3-3ε Protein Western Blot Y75-30N 14-3-3ε
Table S2. Sequences immunoprecipitating with anti-hif-1α
 Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
SUPPLEMENTAL MATERIAL
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 SUPPLEMENTAL MATERIAL Biogenesis of Pro-senescent Microparticles by Endothelial Colony Forming Cells from Premature Neonates is driven
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 SUPPLEMENTAL MATERIAL Biogenesis of Pro-senescent Microparticles by Endothelial Colony Forming Cells from Premature Neonates is driven
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
CD4 + T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2
 Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland
Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland
RHOA inactivation enhances Wnt signaling and promotes colorectal cancer
 RHOA and colorectal cancer Paulo Rodrigues et al 2014 SUPPLEMENTARY INFORMATION RHOA inactivation enhances Wnt signaling and promotes colorectal cancer Paulo Rodrigues, Irati Macaya, Sarah Bazzocco, Rocco
RHOA and colorectal cancer Paulo Rodrigues et al 2014 SUPPLEMENTARY INFORMATION RHOA inactivation enhances Wnt signaling and promotes colorectal cancer Paulo Rodrigues, Irati Macaya, Sarah Bazzocco, Rocco
 Νόσος Parkinson-Νέοι ορίζοντες Ξηροµερήσιου Γεωργία Νευρολόγος-Μέλος ερευνητικής οµάδας Νευρογενετικής Αίτια της νόσου Parkinson Περιβαλλοντικοί παράγοντες Γενετικοί παράγοντες Γενετική βλάβη (παθογόνα
Νόσος Parkinson-Νέοι ορίζοντες Ξηροµερήσιου Γεωργία Νευρολόγος-Μέλος ερευνητικής οµάδας Νευρογενετικής Αίτια της νόσου Parkinson Περιβαλλοντικοί παράγοντες Γενετικοί παράγοντες Γενετική βλάβη (παθογόνα
Analysis Summary: Hypermobility Beighton Score
 Analysis Summary: Hypermobility Beighton Score Phenotype Description The beighton_hypermobility_qtl phenotype comes from: morphology survey: Can you bring your right thumb to reach your forearm (see above
Analysis Summary: Hypermobility Beighton Score Phenotype Description The beighton_hypermobility_qtl phenotype comes from: morphology survey: Can you bring your right thumb to reach your forearm (see above
ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ
 5 ο ΠΑΝΕΛΛΗΝΙΟ ΣΥΝΕΔΡΙΟ ΕΛΛΗΝΙΚΗΣ ΕΤΑΙΡΕΙΑΣ ΑΙΜΑΦΑΙΡΕΣΗΣ ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ Θωμάς Τσαγανός, MD, PhD Πανεπιστημιακός Υπότροφος Δ Παθολογική Κλινική Πανεπιστημιακό Γενικό Νοσοκομείο
5 ο ΠΑΝΕΛΛΗΝΙΟ ΣΥΝΕΔΡΙΟ ΕΛΛΗΝΙΚΗΣ ΕΤΑΙΡΕΙΑΣ ΑΙΜΑΦΑΙΡΕΣΗΣ ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ Θωμάς Τσαγανός, MD, PhD Πανεπιστημιακός Υπότροφος Δ Παθολογική Κλινική Πανεπιστημιακό Γενικό Νοσοκομείο
Systematic Analysis of Candidate Genes for Alzheimer s Disease in a French, Genome-Wide Association Study
 Journal of Alzheimer s Disease 20 (2010) 1 13 1 IOS Press Supplementary Material Systematic Analysis of Candidate Genes for Alzheimer s Disease in a French, Genome-Wide Association Study Geoffroy Laumet
Journal of Alzheimer s Disease 20 (2010) 1 13 1 IOS Press Supplementary Material Systematic Analysis of Candidate Genes for Alzheimer s Disease in a French, Genome-Wide Association Study Geoffroy Laumet
Γενετική Προδιάθεση Ψωρίασης - Πού βρισκόμαστε?
 Γενετική Προδιάθεση Ψωρίασης - Πού βρισκόμαστε? Κατερίνα Πατσατσή Λέκτορας Δερματολογίας Αφροδισιολογίας Β Κλινική Δερματικών & Αφροδισίων Νόσων ΑΠΘ Νοσοκομείο Παπαγεωργίου ΓΕΝΙΚΑ ΣΤΟΙΧΕΙΑ Προσβάλλει 0.2
Γενετική Προδιάθεση Ψωρίασης - Πού βρισκόμαστε? Κατερίνα Πατσατσή Λέκτορας Δερματολογίας Αφροδισιολογίας Β Κλινική Δερματικών & Αφροδισίων Νόσων ΑΠΘ Νοσοκομείο Παπαγεωργίου ΓΕΝΙΚΑ ΣΤΟΙΧΕΙΑ Προσβάλλει 0.2
Optimizing Microwave-assisted Extraction Process for Paprika Red Pigments Using Response Surface Methodology
 2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
Μοριακή βάση του καρκίνου της ουροδόχου κύστεως
 Πανεπιστήμιο Αθηνών Τμήμα Βιολογίας Τομέας Βιοχημείας & Μοριακής Βιολογίας Μοριακή βάση του καρκίνου της ουροδόχου κύστεως Μαργαρίτης Αυγέρης, Ανδρέας Σκορίλας Τομέας Βιοχημείας και Μοριακής Βιολογίας,
Πανεπιστήμιο Αθηνών Τμήμα Βιολογίας Τομέας Βιοχημείας & Μοριακής Βιολογίας Μοριακή βάση του καρκίνου της ουροδόχου κύστεως Μαργαρίτης Αυγέρης, Ανδρέας Σκορίλας Τομέας Βιοχημείας και Μοριακής Βιολογίας,
Differentially expressed genes of HEK293/HeLa Myst4 sirna vs. controls.
 Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
