LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
|
|
|
- Ῥόδη Δουρέντης
- 6 χρόνια πριν
- Προβολές:
Transcript
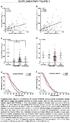
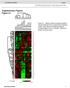
1 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I. Wistuba, Humam Kadara. Supplementary figure legends Supplementary Figure 1. Western blotting of LAPTM4B protein following RNA interference-mediated knockdown. Calu-6 lung cancer cells were transfected with control or LAPTM4B-specific sirnas and protein lysates were isolated and subjected to SDS-PAGE as detailed in the Materials and Methods section. Western blotting analysis was then performed for LAPTM4B protein. Membranes were stained with antibody against β-actin to ensure equal loading of proteins. Supplementary Figure 2. LAPTM4B over-expression increases NRF2 and HMOX1 mrna levels. Calu-6 lung cancer cells were transfected with control or LAPTM4B over-expression vectors. One day following transfection, cells were washed twice with 1x PBS and then incubated in cell culture medium containing 0% or 10% FBS for the indicated time points. Total RNA was isolated from all samples and analyzed for LAPTM4B (upper), NRF2 (middle) and HMOX1 (lower) expression levels by qrt-pcr as detailed in the Materials and Methods section. Expression changes are depicted relative to the first sample (cells transfected with control vector and grown in medium containing 10% FBS for 48 h). qrt-pcr analysis was performed in triplicates for all samples. * indicate P-values < 0.05 assessed by the Student s t-test.
2 Supplementary Figure 3. LAPTM4B knockdown suppresses expression of NRF2 target genes. Calu-6 lung cancer cells were transfected with control, LAPTM4B-specific or NRF2- targeting sirnas. One day following transfection, cells were washed twice with 1x PBS and then incubated in cell culture medium containing 0% FBS for the indicated time points. Total RNA was isolated from all samples and analyzed for SLC7A11 (upper), ME1 (middle) and NQO1 (lower) expression levels by qrt-pcr as detailed in the Materials and Methods section. Expression changes are depicted relative to the first sample (cells transfected with control sirna. qrt-pcr analysis was performed in triplicates for all samples. * indicate P-values < 0.05 assessed by the Student s t-test. Supplementary Figure 4. LAPTM4B over-expression increases NRF2 nuclear localization. Calu-6 lung cancer cells were transfected with control or LAPTM4B over-expressing vectors. One day following transfection, cells were incubated in culture medium containing 0% or 10% FBS for the indicated time points after which total and nuclear protein lysates were isolated and subjected to SDS-PAGE as detailed in the Materials and Methods section. Western blotting analysis was then performed for total and nuclear NRF2 protein levels. Membranes were stained with antibodies against Fibrillarin and β-actin to ensure equal loading of nuclear and total proteins, respectively.
3 Supplementary figure 1 sicontrol: + - silaptm4b: - +
4 Relative expression Supplementary figure LAPTM4B * * * NRF2 * * * * 650 HMOX * 2 * 0 10% FBS: Control vector: LAPTM4B: * * 48 h 72 h
5 Relative expression Supplementary figure * SLC7A * * * * ME * * * * NQO * * * 0 sicontrol silaptm4b sinrf2 sicontrol silaptm4b sinrf2 48 h 72 h
6 Supplementary figure 4 6 h 24 h 10% FBS: Control vector: LAPTM4B: NRF2 Fibrillarin Nuclear NRF2 b-actin Total
7 Supplementary Table 1. Clinicopathological information of the NSCLC tissue microarray specimens analyzed by in situ hybridization Covariate Covariate categories Frequency (percentage) Race African American 17 (4.6%) Asian 5 (1.4%) Hispanic 10 (2.7%) Caucasian 336 (91.3%) Gender Male 174 (47.3%) Female 194 (52.7%) Tobacco history No 43 (11.7%) Yes 325 (88.3%) Smoking status Never 43 (11.7%) Former 173 (47.0%) Current 152 (41.3%) Histology Adenocarcinoma 245 (66.6%) Squamous cell carcinoma 123 (33.4%) Grade Well 45 (12.2%) Moderate 200 (54.4%) Poor 123 (33.4%) Stage I 224 (60.9%) II 63 (17.1%) III 66 (17.9%) IV 15 (4.1%)
8 Supplementary Table 2. Mean LAPTM4B in situ hybridization score in lung adenocarcinomas and squamous cell carcinomas Histology Mean ISH score LUAD 22.8 SCC 18.4 ISH, in situ hybridization; LUAD, lung adenocarcinoma; SCC, squamous cell carcinoma
9 Supplementary Table 3. Gene features significantly differentially expressed in serum starved lung cancer cells transfected with LAPTM4B -specific sirna compared to cells transfected with control sirna Up-regulated gene features following sirna-mediated LAPTM4B knockdown Probeset Accession Symbol p-value NM_ SPON NM_ ADAM NM_ NTSR NM_ FRMD NM_ LAMA NM_ RNF NM_ ARHGAP NM_ KLRC NM_ PIWIL NM_ RAB3B NM_ SIX NM_ TNC NM_ HBA NM_ HBA NM_ SVIP NM_ ACP NM_ PLEKHH NM_ PLAU NM_ IL17RD NM_ BCAM NM_ SOX NM_ LAMC NM_ ANKRD NM_ NEBL NM_ FAM55B NM_ DCLK NM_ ULBP NM_ C9orf NM_ RNF NM_ WDFY NM_ FLRT NM_ SEMA3E NM_ TFRC NM_ SIGIRR NM_ IGFBP NM_ PAX NM_ CTHRC NM_ IL31RA NM_ CDYL NM_ JAG NR_ LINC NM_ PPP2R1B NM_ MORC NM_ NAV
10 NM_ CCDC NM_ ECM NM_ KCNJ NM_ DSP NM_ ZNF75A NM_ OLFML NM_ ZBTB NM_ C11orf NM_ CRIP ENST NM_ HMMR NM_ HSPG NM_ LMNB NM_ FLNB NM_ LAMA NM_ MYB NM_ DAB NM_ GPRC5A NM_ CCDC NM_ DPYSL ENST NM_ KIAA NM_ SASH NM_ KLRC NM_ NAA NM_ MORC NM_ ADRA2C NM_ BMP NM_ MYH NM_ CARD NM_ CHST NM_ BAIAP NM_ TEX NM_ CRYZ NM_ CHD NM_ HIPK NR_ MIR181A ENST NM_ ZBTB NM_ MALL NM_ ASAP NM_ RGL NM_ FBXL NM_ ANKRD AK FLJ NM_ KLRC NM_ SYNE NM_ SULF NM_ PSD NM_ HS3ST
11 ENST ENST ENST NM_ UNC5A NM_ ITGA NM_ ITGA NM_ ACCN NM_ GEMIN NM_ KLRK NM_ RGL NM_ RGL NM_ NOG NM_ TP53INP NM_ ACVR2B NM_ PTP4A ENST NM_ ZNF ENST NM_ SLC2A ENST NM_ LSP NM_ HOXB NM_ TEAD NM_ TXLNA NM_ C11orf NM_ CEACAM NM_ EOMES NR_ SNORA71C ENST NM_ C19orf NM_ RNH NM_ BMP2K NM_ GGT NM_ LAT NM_ CCDC ENST NM_ TRAM ENST NM_ ZNF NM_ FURIN NM_ EPHB NM_ PLXNA NM_ B3GALT BC NM_ CMTM ENST NM_ VPS13A ENST ENST NM_ SLIT NM_ EPHB NM_ CAB39L NM_ ARHGAP
12 NM_ TMC NM_ ARRB NM_ NDRG NM_ FBXL NM_ REEP AK NR_ SMEK3P NM_ LAMB NM_ MIF4GD NM_ STC NM_ ZFP NM_ RHPN NM_ CDC42EP NM_ MTSS NM_ HMGA NM_ C14orf NM_ PLCH NM_ SLC16A NM_ APOBEC3G NM_ IL NM_ CACNA1C NM_ DNAH ENST ENST NM_ SYCP ENST NM_ AGA NM_ FAM36A ENST NM_ ACSS NM_ KRT NM_ FGF NM_ BHLHB NM_ CD NM_ MAP4K ENST NM_ ZNF NM_ ENDOV NM_ ARFIP NM_ ZNF NM_ PELO
13 NM_ SFXN ENST NM_ CHD NM_ GIPC NM_ COL13A NM_ DCBLD NM_ Mar NM_ PRKCDBP NM_ TET NM_ C10orf ENST NM_ AMOTL ENST NR_ FBXL19-AS NM_ COMMD NR_ SNORA NM_ GPR NM_ RASGRF NR_ NBPF22P NM_ LRRC NM_ CITED NM_ EPHA NM_ ANTXR NM_ ZNF NM_ NFIA ENST NM_ MCTP NM_ RALGDS NM_ CD3EAP NM_ LAPTM NM_ TMEM NM_ TMEM132C NM_ SGPL NM_ PLK NM_ MASP NM_ ARSD ENST NR_ TERC NM_ KIF NM_ NLRP NM_ CDR2L NM_ CD NM_ FAM102B ENST ENST NM_ IDH ENST NM_ KIFAP
14 NM_ VEZF NM_ PDGFRA NM_ LRRC NM_ AKNAD NM_ GLI NM_ SERPINA NM_ NCEH AF NM_ NAPRT NM_ MMP NM_ DDAH NM_ DDAH ENST NM_ FAAH NM_ ABCB NR_ GGT3P ENST NR_ FAM27A NM_ PRIC NM_ MPI NM_ SERTAD NM_ HMGCS NM_ CGNL NM_ CDH NM_ SPATA NM_ APBB NM_ MLPH NM_ CELSR NM_ C17orf NM_ SH3TC NM_ SLC26A ENST NM_ USP NM_ ATP8B NM_ ESRRB NM_ DOCK NR_ FAM27C NM_ TM4SF NR_ FAM27A NM_ HEY NM_ SKAP AK NM_ RAB3D NR_ LOC NM_ ACBD ENST NM_ CRIP ENST
15 NM_ DCP NM_ NUAK NM_ AGAP NM_ CACNG NM_ AGRN NM_ BMP8A NM_ CNN NM_ EIF2C NM_ KRT NM_ DAB2IP NM_ RAET1E NM_ FAM200B ENST NM_ C3AR NM_ ENTPD NM_ KLHL NM_ ANKRD AK LOC NM_ SRPX NM_ ARHGEF NM_ ESR NM_ MALT NM_ KRTAP NM_ C5orf NR_ FLJ NM_ FAM48B NM_ FAM48B NM_ PKN NM_ ADAMTS NM_ GIN ENST NM_ NUMA NM_ WNK NM_ ASTN NM_ SPDYE NM_ NUDT NM_ DDAH NM_ PCMTD ENST NM_ HIST1H2BJ NM_ NEB NM_ ZNF NM_ ETS NM_ SLCO4A ENST NM_ APOBEC3B NM_ LBP
16 NM_ FBXL NM_ NARF NM_ SPTB ENST NM_ REEP NM_ CACNA1H NM_ SPNS ENST NR_ GGT3P NM_ SCLT NR_ SNORA71A NM_ FAM55D NM_ GCC NR_ FUNDC2P NM_ KCTD NM_ MIA NM_ SIRT NM_ ITGAM NM_ RRNAD NM_ CXCL NM_ DLL NM_ GRB NM_ KLRC NM_ RHBG NM_ NM_ SH2D4A AY NSAP NM_ CDKN2AIPNL NM_ MBD3L NM_ AKAP NM_ ZNF NM_ NOX AK LOC AF ENST NM_ FSCN NM_ MORC Down-regulated gene features following sirna-mediated LAPTM4B knockdown Probeset Accession Symbol p-value NM_ LAPTM4B NM_ SLC1A NM_ SPP NM_ AK NM_ IL7R NM_ PDZK
17 NM_ SLC38A NM_ MGLL NM_ SEL1L NM_ SEMA3C NM_ PTGS NM_ SLC7A NM_ SLC35B NM_ GCNT NM_ SLC7A NM_ DENND2D NM_ ACTG NM_ DHRS NM_ DAPK NM_ TIMP NM_ ANK NM_ TBXAS NM_ CDKN1A NM_ PCTP NM_ CYP1B NM_ SRPX NM_ ADCY NM_ ADIPOR NM_ FAM171B NM_ RDH NM_ FGF NM_ AKR1C NM_ EGR NM_ RGS NM_ ELMO NR_ NM_ F2RL NM_ LYPD6B NM_ VWDE NM_ PPDPF NM_ CFH NM_ PHTF NM_ REN NM_ JPH NM_ CD NM_ PLA2G AL NM_ CXCL NM_ HMOX NM_ PIP5K1B NM_ PPAP2A NM_ LIMA NM_ ME NM_ ATP10D NM_ ROR NR_ MIR22HG NM_ DSEL NM_ PTPRE NM_ KRT NM_ GEM
18 NM_ HS6ST NM_ BPNT NM_ ARAP NM_ SLITRK NM_ FFAR NM_ CHRNA NM_ FABP NM_ SLC14A NM_ CA5B NM_ PAPPA NM_ GABRR NM_ PLA2G4A NM_ SLC4A ENST NM_ GXYLT NM_ CPE NM_ G6PD NM_ SNAI NM_ GPR NM_ B4GALT NM_ CREB3L NM_ GNPDA NM_ MCTP NM_ RUNX NM_ ADORA2B NM_ B3GALNT NM_ NQO NM_ AIG NM_ FOSL NM_ LOX NM_ FZD NM_ TMEM NM_ PPP2R3A NM_ MAL NM_ LPXN NM_ IL1RAP NM_ NDST NM_ MMD NM_ SNCA NM_ CD NM_ TFF NM_ TMEM NM_ PRCP NM_ SCAMP NM_ SGK NM_ HTATIP NM_ PLA2G NM_ ODZ NM_ MFAP
19 NM_ CKAP NM_ RAPGEF NM_ GPR NM_ BCL2L NM_ LIMCH NM_ HPGD NM_ KCNH NM_ GPR NM_ GBP NM_ PPM1L NM_ RGAG NM_ HSPA NM_ PLIN NM_ CNIH NM_ ARHGEF NM_ THBS NM_ FOS NM_ TRIM NM_ CYBRD NM_ GBP NM_ FGFR NM_ NDEL NM_ SLC41A NM_ SULT1C NM_ RBPMS NM_ TIMP NM_ PGBD NM_ PDGFRB NM_ PAMR NM_ SLC16A NM_ IL6ST NM_ PCDHB NM_ SLFN NM_ GDF NM_ HMGCS NM_ FBN NM_ ACVR NM_ GPRC5B NM_ CDCA7L NM_ MTUS NM_ ABCC NM_ DUSP NM_ PIR NM_ POPDC NR_ MIR NM_ FAM26E NM_ FAM55C AK FSIP NM_ TRIM16L NR_ HHATL NM_ SEZ NM_ LRRC NM_ CYB5R NM_ NR4A NM_ SLC44A
20 NM_ B4GALNT NM_ Sep NM_ PTPN NM_ PION NM_ IDS NM_ ATL NM_ ARHGAP NM_ GNG NM_ SHANK NM_ COG NM_ ABCC AY C6orf NM_ PDE1B NM_ TRAPPC NM_ ABCA NM_ GAB NM_ NFKBIZ NM_ ACAD NM_ GABRE NM_ KCTD NM_ AVPI NM_ CCNE NM_ ENTPD NM_ TMTC NM_ PPM1K NM_ MB21D NM_ GCLM NM_ QPRT NM_ TFF NM_ ELMO BC C11orf NM_ C1orf NM_ MTERFD NM_ ASB NM_ DNAJB NM_ SLC37A NM_ LUM NM_ SLC29A NM_ OCRL NM_ CD NM_ IDS NM_ CD NM_ IFITM NM_ DUSP NM_ PTPRN NM_ TMEM167A NM_ RGS NM_ SOCS ENST NM_ KCNN NM_ FRY
21 NM_ FNDC NM_ SNX NM_ AKAP NM_ LNX NM_ C7orf NM_ UCHL NM_ MXD NM_ LIF NM_ HIP1R NM_ CPNE NM_ PTGER NM_ L1CAM NM_ APOL NM_ CADPS NR_ MGC NM_ VWDE NM_ AMZ NM_ BHLHE NM_ PLCE NM_ PIAS NM_ UST NM_ CDHR NM_ S100A NM_ ANGPTL NR_ MIR NM_ ATXN7L BC C12orf NM_ PTPN NM_ DLX NM_ ABHD NM_ TXLNG NM_ PHEX NM_ PRSS NM_ LONRF NM_ CERS NR_ MIR NM_ PLEKHA NM_ RAB NM_ SRGN NM_ SRM NM_ BMP NM_ PLXDC NM_ F13A NM_ SEMA6A NM_ INSIG NM_ GATA NR_ C14orf NM_ PIM NM_ HECTD NM_ CEACAM NM_ RAB2B NM_ TGFBRAP NM_ PARM NM_ GLRA NM_ ADAM
22 NM_ SH3BP NM_ TMEM106C NR_ BRP NM_ LARGE NM_ DISP NM_ MTCP NM_ PRKG NM_ AHCYL NM_ LCP NM_ HSPA NM_ PHF NM_ DDX NM_ FAM126B NM_ RAB8B NM_ TFF NM_ UBL NM_ TMED NM_ CHGA NM_ DLX NM_ FAM59A NM_ PAG NM_ SLC16A NM_ MAPRE NM_ RIMKLB NM_ DNAJA NM_ MFSD NM_ PARP NM_ RPIA NM_ WDFY NM_ RNF NM_ IL NM_ ADM NM_ ANO NM_ FZD NM_ PTPN NM_ SNX NM_ CHRM NM_ ANG NM_ NQO NM_ SLC25A NM_ TMEM167B NM_ TMEM167B NM_ THRB NM_ NAA NM_ KCNK NM_ EFNA NM_ DRD NM_ TRPC NM_ SPARC
23 NM_ CACNA2D NM_ NLRP NM_ SLC4A NM_ RGL NM_ PLAGL NM_ MMP NM_ ADRBK NM_ CHMP2B NM_ TMEM NM_ MCOLN NM_ TMTC NM_ VSIG10L NM_ AMIGO NM_ DEGS NM_ HOMER NM_ DHDDS NR_ KGFLP NM_ TPD52L NM_ RASGEF1B NM_ GSTK NM_ UGDH NM_ B3GNT NM_ BAD NM_ TSKU NM_ PION NM_ CCPG NM_ AKR1C NM_ DCAF NM_ SLC9A NM_ C7orf NM_ LRRC8C NM_ NR3C NM_ ADAM NM_ MAPK NR_ MIR31HG NM_ BMP NM_ FAM122B NM_ CKMT1A NM_ CKMT1A NM_ TOM1L NM_ NARS NM_ STXBP5L NM_ C6orf NM_ C6orf NM_ CDH NM_ WIPI NM_ TTLL
24 NM_ ID NM_ ATP6V0E NM_ AKAP NM_ CDH NM_ TBC1D NM_ MAOA NM_ NNT NM_ DPYD ENST AK NM_ EFHD NM_ RILPL NM_ EIF4EBP NR_ RPSAP NM_ LHFP NM_ TSPAN NM_ SMPD NM_ SLC4A NM_ FSTL NM_ SERPINH NM_ CRY NM_ RET AK FSIP NM_ WFS NM_ POR NM_ COBLL NM_ ANGPTL NM_ TMEM63C NM_ MFSD2A NM_ PDE4DIP NM_ CASP NM_ RHOQ NM_ NCAM NM_ LGR NM_ OVGP NM_ SFXN NM_ PTPRN NM_ VSTM2L NM_ PRKCZ NM_ CASD NM_ CDK NM_ ANKRD36B NR_ SNORD NM_ FAM134B NM_ S100P ENST NM_ SPIRE NM_ SPATA NM_ ERG NM_ PTX NM_ ECHDC
25 NM_ C7orf NM_ MANBA NM_ CYTH NM_ NR4A NM_ ALDH3A NM_ RYBP NM_ IGFBP NM_ RGS NM_ MYO NM_ RFX NM_ RPS NM_ RABGGTB NM_ RAB3C NM_ PYGL NM_ TRAK NM_ ABCC NM_ ZDHHC NM_ SMYD NM_ BST NM_ PCDH NM_ TMEM126A NM_ MBL NM_ CNTNAP NM_ KHDRBS NM_ UBE2W NM_ SH3BGRL NM_ LPHN NM_ CD99L NM_ SEMA5A NR_ SNORD14C NM_ PROS NM_ B3GAT NM_ FERMT NM_ CSRNP NM_ TXNRD NM_ AKR1C NM_ SEC61A NM_ GNPDA NM_ ANGPT NM_ APBB NM_ ZNF NM_ C14orf NM_ EGR NM_ OSTF NM_ C9orf NM_ CFHR BC C16orf NM_ ZFP NM_ FAM188A
26 NM_ SLC22A NM_ CDK NM_ CDK NM_ GDE NM_ LRIG NM_ NFKBID NM_ HSD17B NR_ CATSPER2P AY DDR NM_ SSH NM_ TIMP NM_ RECK NM_ SMAP NM_ HLCS NM_ FAM49A ENST NM_ UCP NM_ KIAA NM_ TEX NM_ WDR NM_ SPECC NM_ SNTB NM_ TPMT NM_ CA NM_ EFEMP NM_ EMP NM_ BOC NM_ ABHD NM_ MCF NM_ C19orf NM_ SCPEP NM_ LRRTM NM_ OASL NM_ FAH NM_ LGI NM_ ZNF NM_ FTL NM_ PRAME NM_ MATN NM_ DNM NM_ CLN NM_ AREG NM_ SRXN NM_ DUSP GENSCAN NM_ RAB NM_ NSMCE4A NM_ SLC25A NM_ PLAC
27 NR_ LGALS NM_ GOLIM NM_ SLC20A NM_ FBXO NM_ SESN NM_ CLN NM_ DNER NM_ GPR NM_ MOSC NM_ ACCN NM_ CCDC109B NM_ DLL NM_ GNA NM_ POF1B NM_ KIAA ENST NM_ SLC25A NM_ DRD NM_ C6orf ENST NM_ MLKL NM_ QDPR NM_ TGFBR NR_ KGFLP NR_ KGFLP NR_ KGFLP NM_ CPD NM_ SEC24D NM_ IFNAR NM_ TGFB NM_ KIAA NR_ TRAF3IP NM_ C6orf NM_ BMP NM_ SERINC NM_ FGL NM_ STEAP NM_ NADSYN NM_ ABHD NM_ RPL37A NM_ FKBP NM_ HOXA NM_ KCNS NM_ PRR NM_ TYRO NM_ GALNT
28 NM_ PNOC NM_ NEK AY SNHG NM_ E2F NM_ NR1H NM_ CEBPB NM_ GSTM NM_ SLC29A NM_ GOLGA NM_ ARNTL NM_ SYT NM_ FITM NM_ GPD1L NM_ FEZ NM_ SQSTM NM_ ABCA NM_ SOD NM_ DNAJC NM_ GCLC NM_ SDR16C NM_ F11R NM_ LPAR NM_ ISOC NM_ PPP3CA NM_ AGPAT NM_ RTN BX NM_ CPLX NR_ DLEU NM_ RWDD NM_ CDC14B NM_ RNF NM_ CLIP NM_ WDR NM_ GTF3C NM_ CDK NM_ FAM40B NM_ DSE NM_ FSTL NM_ VPS NM_ FYN NM_ ARSA NM_ NEU NM_ NEU NM_ ZDHHC NR_ LINC NM_ FBXO
29 NM_ LRP5L NM_ ADPRH NM_ C9orf NM_ IQCG NM_ LYZ NR_ AMZ2P NM_ ABHD NM_ CNKSR NM_ TSPAN NM_ GTF2E NM_ IQCK NM_ PIK3CA NM_ UAP NM_ CASK NR_ LOC NM_ HMCN NM_ ABCG NM_ POLR1D NM_ HAUS NM_ DNAJB NM_ PRICKLE NM_ GOLGA7B NM_ POFUT NM_ GPAA NM_ TOX NM_ PLA2R NM_ TMEM NM_ NIPAL NM_ LEPREL NM_ PGCP NM_ INTS NM_ STRA NM_ NEU NM_ MBOAT NM_ PKDCC NM_ COQ10A NM_ MBNL NM_ PNP NM_ SEPW NM_ NRIP NM_ KLHL NM_ TMEM NR_ C14orf NM_ C5orf NM_ FTH NM_ TSPAN NM_ NCOA NM_ EIF2B NM_ CLIP
30 NM_ C18orf NM_ PAK NM_ SYBU NM_ NTN NM_ FAM111A NM_ JHDM1D AK LOC AY NM_ ATP1B AK LOC NM_ MMP NM_ GLCCI NM_ USP NR_ SNORD NM_ TMEM229B NM_ IFI NM_ VEZT NM_ TIAM NM_ RBM NM_ CCL NM_ SGIP NM_ TSPYL NM_ C11orf NM_ OSBPL1A NM_ VANGL NM_ FBXO NM_ SGTB NM_ AS3MT NM_ RNF NM_ ANUBL NM_ CACNG NM_ NM_ SLC35D NM_ CCDC NM_ SETD NM_ MTX ENST NM_ RNF NM_ EPS NM_ BDH NM_ CDO NM_ NSMAF NM_ ZDHHC NM_ MANSC NM_ ELOVL NM_ B4GALT NM_ ACP NM_ NLN NM_ GBP NM_ ARNT
31 NM_ SCMH NM_ FUT NM_ GPR NM_ OXGR NM_ TSC22D NM_ IGSF NM_ DEPTOR NM_ MAPT NM_ GSS NM_ PTPRJ NM_ GPX NM_ HBE NM_ PXK NM_ PPP1R13B NM_ TBC1D NM_ NUCB NM_ GAB NM_ OBFC2A NM_ WDPCP NM_ CPEB NM_ EPM2A NM_ SLC40A NM_ HENMT NM_ SYNJ NM_ UHMK NM_ SLC25A NR_ LOC NM_ TRMT2B NM_ OAS NM_ USP27X NM_ ACER NM_ TSPAN NM_ FAM190A NM_ ABCB NM_ HGSNAT NM_ FRRS NM_ APH1B NM_ EREG NM_ C3orf NM_ PPARGC1A NM_ TMEM38B NM_ RPL27A
32 Supplementary Table 4. Gene features significantly differentially expressed in lung cancer cells cultured in medium containing 10% FBS and transfected with LAPTM4B - specific sirna compared to cells transfected with control sirna Up-regulated gene features following sirna-mediated LAPTM4B knockdown Probeset Accession Symbol p-value NM_ PRRX NM_ LAMA NM_ IL17RD NM_ ADAM NM_ ANKRD NM_ HAS NM_ RNF NM_ NDRG NM_ DCLK NM_ ARHGAP NM_ THSD7A NM_ NTSR NM_ IL NM_ WDFY NM_ FRMD NM_ KIAA NM_ KLRC NM_ NM_ MPZL NM_ SPON NM_ SEMA3E NM_ ACP NR_ LINC NM_ MORC NM_ TM4SF NM_ SVIP NM_ PMEPA NM_ FST NM_ CGNL NM_ FAM102B NM_ SRPX NM_ KLRC NM_ KLRC NM_ INHBA NM_ PARP NM_ ACSL NM_ MMP NM_ LMNB NM_ SHC NM_ ITGA NM_ ARHGAP NM_ LRRC NM_ TMCC NM_ MAPK NM_ NEK NM_ TMC NM_ MORC
33 NM_ NUDT NM_ HBA NM_ HBA NM_ NR5A NM_ EPCAM NM_ PPP2R1B NM_ ST6GALNAC NM_ ST6GALNAC NM_ MALL NM_ ANTXR NM_ SULF NM_ PIK3AP NM_ C9orf NM_ PLEK NM_ LAPTM NM_ GPR NM_ IFI NM_ CD NM_ PAX NM_ SYPL AF NM_ TCN NM_ SYNM NM_ EPHB NM_ CYP7A NM_ C11orf NM_ OR4C NM_ PDE8B NM_ DDX60L NM_ HIPK NM_ MTAP ENST NM_ CCDC NM_ AHNAK NM_ KLHL NM_ MYB NM_ L3MBTL NM_ FGF NM_ SH3TC NM_ TMTC ENST NM_ LAMC NM_ KIF NM_ ABLIM NM_ SPTB NM_ B3GALT NM_ TXLNA NM_ FBXL NR_ UCA NM_ ATG NM_ FRMD
34 NM_ FILIP1L NM_ REEP NM_ CARD NM_ PAM NM_ SLK NM_ SUCLG NM_ ARHGAP NM_ ST6GALNAC NM_ ITPR NM_ C11orf NM_ IRAK NM_ ASAP NM_ MYH NM_ KIAA NM_ SSFA NM_ ZNF NM_ UNC13B NM_ ADAM NM_ TRAM NM_ KBTBD NM_ CIT NM_ ANKRD NM_ DGKD NM_ NIPAL BC FAM27E NM_ HIST1H2BK NM_ CLDN NM_ TNFRSF BC FAM27E NM_ BIRC BC FAM27E NR_ SNORA38B NR_ TTLL NM_ APOBEC3B NM_ CCDC NM_ CYP1A NM_ DOCK NM_ KLHL NM_ WRAP NM_ NM_ OSGEPL NM_ CD NM_ SPRY NM_ IGFBP NM_ SUSD NM_ CADPS NM_ FKBP NM_ FAM160B NM_ REEP NM_ PLEKHH NM_ MFSD
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Nature Medicine doi: /nm.2772
 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent
 Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT
 Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
 Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis
 S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells
 Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
Supplemental Information. A p53 Super-tumor Suppressor Reveals a Tumor. Suppressive p53-ptpn14-yap Axis. in Pancreatic Cancer
 Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Information for. RRM2B Suppresses Activation of the Oxidative Stress Pathway and is. Up-regulated by P53 During Senescence
 Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
NCBI ID Symbol 2 A2M A4GALT A4GNT 8086 AAAS 13 AADAC AADACL AAGAB 15 AANAT 16 AARS AARS AASS 18 ABAT 19 ABCA1
 NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in. and *Christina Chan
 Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
500 New CUSABIO Antibodies at MedStore!
 BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
Supplementary Table S1 Demographic characteristics of HCC patients
 Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Zhang et al., Supplemental figures, Methods, References and Tables
 1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
Supplementary Figure 1.
 Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Materials for
 www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary data to: The mir-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2
 Supplementary data to: The mir-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB and ZEB2 Sun-Mi Park, Arti B. Gaur 3, Ernst Lengyel 2 and Marcus
Supplementary data to: The mir-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB and ZEB2 Sun-Mi Park, Arti B. Gaur 3, Ernst Lengyel 2 and Marcus
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
Supplemental file 3. All 306 mapped IDs collected by IPA program. Supplemental file 6. The functions and main focused genes in each network.
 LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
Supplementary Online Content
 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal
 Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Burden of Disease Variants in Participants of the Long Life Family Study. Supplemental Online Material
 Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein
 1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Supplementary Information
 Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary Materials: A Preliminary Link between Hydroxylated Metabolites of Polychlorinated Biphenyls and Free Thyroxin in Humans
 S1 of S11 Supplementary Materials: A Preliminary Link between Hydroxylated Metabolites of Polychlorinated Biphenyls and Free Thyroxin in Humans Eveline Dirinck, Alin C. Dirtu, Govindan Malarvannan, Adrian
S1 of S11 Supplementary Materials: A Preliminary Link between Hydroxylated Metabolites of Polychlorinated Biphenyls and Free Thyroxin in Humans Eveline Dirinck, Alin C. Dirtu, Govindan Malarvannan, Adrian
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Comparative transcriptome analysis of peripheral blood mononuclear cells in
 Comparative transcriptome analysis of peripheral blood mononuclear cells in hepatitis B-related acute-on-chronic liver failure Qian Zhou, Wenchao Ding, Longyan Jiang, Jiaojiao Xin, Tianzhou Wu, Dongyan
Comparative transcriptome analysis of peripheral blood mononuclear cells in hepatitis B-related acute-on-chronic liver failure Qian Zhou, Wenchao Ding, Longyan Jiang, Jiaojiao Xin, Tianzhou Wu, Dongyan
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
Supplementary Materials for
 www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
SUPPLEMENTAL MATERIAL
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 SUPPLEMENTAL MATERIAL Biogenesis of Pro-senescent Microparticles by Endothelial Colony Forming Cells from Premature Neonates is driven
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 SUPPLEMENTAL MATERIAL Biogenesis of Pro-senescent Microparticles by Endothelial Colony Forming Cells from Premature Neonates is driven
Supplementary Table 1. Composition of the diets
 Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in
 1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
Potential Dividers. 46 minutes. 46 marks. Page 1 of 11
 Potential Dividers 46 minutes 46 marks Page 1 of 11 Q1. In the circuit shown in the figure below, the battery, of negligible internal resistance, has an emf of 30 V. The pd across the lamp is 6.0 V and
Potential Dividers 46 minutes 46 marks Page 1 of 11 Q1. In the circuit shown in the figure below, the battery, of negligible internal resistance, has an emf of 30 V. The pd across the lamp is 6.0 V and
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
http / / cjbmb. bjmu. edu. cn Chinese Journal of Biochemistry and Molecular Biology RT-PCR Runx d PCR RT-qPCR Western BMPs Wnt Ihh
 ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 6 27 6 540 ~ 547 micrornas * / / 250062 microrna mrna β- RT-PCR Runx2 0 7 14 d micrornas
ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 6 27 6 540 ~ 547 micrornas * / / 250062 microrna mrna β- RT-PCR Runx2 0 7 14 d micrornas
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion
 Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Η ΥΠΕΡΕΚΦΡΑΣΗ ΤΟΥ SMAD7 ΠΡΟΣΤΑΤΕΥΕΙ ΤΟ ΗΠΑΡ ΑΠΟ ΤΗΝ TGF-Β/SMAD ΜΕΣΟΛΑΒΟΥΜΕΝΗ ΙΝΟΓΕΝΕΣΗ
 Η ΥΠΕΡΕΚΦΡΑΣΗ ΤΟΥ SMAD7 ΠΡΟΣΤΑΤΕΥΕΙ ΤΟ ΗΠΑΡ ΑΠΟ ΤΗΝ TGF-Β/SMAD ΜΕΣΟΛΑΒΟΥΜΕΝΗ ΙΝΟΓΕΝΕΣΗ OVEREXPRESSION OF SMAD7 PROTECTS LIVER FROM TGF-B/SMAD-MEDIATED FIBROGENESIS Γ. Γερμανίδης(1), Ν. Αργέντου(2), Ε.
Η ΥΠΕΡΕΚΦΡΑΣΗ ΤΟΥ SMAD7 ΠΡΟΣΤΑΤΕΥΕΙ ΤΟ ΗΠΑΡ ΑΠΟ ΤΗΝ TGF-Β/SMAD ΜΕΣΟΛΑΒΟΥΜΕΝΗ ΙΝΟΓΕΝΕΣΗ OVEREXPRESSION OF SMAD7 PROTECTS LIVER FROM TGF-B/SMAD-MEDIATED FIBROGENESIS Γ. Γερμανίδης(1), Ν. Αργέντου(2), Ε.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
5- CACGAAACTACCTTCAACTCC-3 beta actin-r 5- CATACTCCTGCTTGCTGATC-3 GAPDH-F GAPDH-R
 Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 651 APPENDIX B. BIBLIOGRAPHY 677 APPENDIX C. ANSWERS TO SELECTED EXERCISES 679
 APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
Heterobimetallic Pd-Sn Catalysis: Michael Addition. Reaction with C-, N-, O-, S- Nucleophiles and In-situ. Diagnostics
 Supporting Information (SI) Heterobimetallic Pd-Sn Catalysis: Michael Addition Reaction with C-, N-, -, S- Nucleophiles and In-situ Diagnostics Debjit Das, a Sanjay Pratihar a,b and Sujit Roy c * a rganometallics
Supporting Information (SI) Heterobimetallic Pd-Sn Catalysis: Michael Addition Reaction with C-, N-, -, S- Nucleophiles and In-situ Diagnostics Debjit Das, a Sanjay Pratihar a,b and Sujit Roy c * a rganometallics
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Supporting Information
 Supporting Information Mitochondria-Targeting Polydopamine Nanocomposites as Chemophotothermal Therapeutics for Cancer Zhuo Wang *,, Yuzhi Chen, Hui Zhang, Yawen Li, Yufan Ma, Jia Huang, Xiaolei Liu, Fang
Supporting Information Mitochondria-Targeting Polydopamine Nanocomposites as Chemophotothermal Therapeutics for Cancer Zhuo Wang *,, Yuzhi Chen, Hui Zhang, Yawen Li, Yufan Ma, Jia Huang, Xiaolei Liu, Fang
Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo
 Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
Original Article Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells
 Int J Clin Exp Med 2014;7(11):4253-4259 www.ijcem.com /ISSN:1940-5901/IJCEM0002259 Original Article Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells Chen Chen 1, Yachun
Int J Clin Exp Med 2014;7(11):4253-4259 www.ijcem.com /ISSN:1940-5901/IJCEM0002259 Original Article Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells Chen Chen 1, Yachun
SUPPLEMENTARY INFORMATION RECURRENT MUTATIONS IN THE U2AF1 SPLICING FACTOR IN MYELODYSPLASTIC SYNDROMES
 SUPPLEMENTARY INFORMATION RECURRENT MUTATIONS IN THE U2AF1 SPLICING FACTOR IN MYELODYSPLASTIC SYNDROMES Timothy A Graubert 1 3,9, Dong Shen 4,9, Li Ding 4,5,9, Theresa Okeyo-Owuor 1, Cara L Lunn 1, Jin
SUPPLEMENTARY INFORMATION RECURRENT MUTATIONS IN THE U2AF1 SPLICING FACTOR IN MYELODYSPLASTIC SYNDROMES Timothy A Graubert 1 3,9, Dong Shen 4,9, Li Ding 4,5,9, Theresa Okeyo-Owuor 1, Cara L Lunn 1, Jin
Nature Immunology: doi: /ni Supplementary Figure 1. Foxm1 loss leads to bone marrow hypocellularity.
 Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver
 Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Kansas Culture Change Instrument KCCI. resident centered care, a homelike environment, staff/resident relationships, staff
 73 KansasCultureChangeInstrumentKCCI residentcenteredcare,ahomelikeenvironment,staff/residentrelationships,staff empowerment,nursinghomeleadershipsqualityimprovement sharevalues nkcci. rp 74 75 . ProblemandPurpose
73 KansasCultureChangeInstrumentKCCI residentcenteredcare,ahomelikeenvironment,staff/residentrelationships,staff empowerment,nursinghomeleadershipsqualityimprovement sharevalues nkcci. rp 74 75 . ProblemandPurpose
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
J. Dairy Sci. 93: doi: /jds American Dairy Science Association, 2010.
 Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros
SUPPLEMENTARY INFORMATION doi:10.1038/nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros
Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών
 Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Analysis Summary: Any Allergy
 Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Differentially expressed genes of HEK293/HeLa Myst4 sirna vs. controls.
 Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
