CD4 + T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2
|
|
|
- Τίμω Κύνθια Λύκος
- 6 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland Tschismarov 1,11, Lisa Göschl 1,2, Mirjam A Moser 3, Sabine Lagger 3,10, Shinya Sakaguchi 1, Mircea Winter 3, Florian Lenz 4, Dijana Vitko 5, Florian P Breitwieser 5, Lena Müller 1, Hammad Hassan 1, Keiryn L Bennett 5, Jacques Colinge 5, Wolfgang Schreiner 4, Takeshi Egawa 6, Ichiro Taniuchi 7, Patrick Matthias 8,9, Christian Seiser 3,12 & Wilfried Ellmeier 1,12 1 Division of Immunobiology, Institute of Immunology, Center for Pathophysiology, Infectiology and Immunology, Medical University of Vienna, Vienna, Austria. 2 Division of Rheumatology, Medicine III, Medical University of Vienna, Vienna, Austria. 3 Department of Medical Biochemistry, Max F. Perutz Laboratories, Vienna Biocenter, Medical University of Vienna, Vienna, Austria. 4 Division of Biosimulation and Bioinformatics, Center for Medical Statistics, Informatics, and Intelligent Systems, Medical University of Vienna, Vienna, Austria. 5 CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria. 6 Department of Pathology & Immunology, Washington University School of Medicine, St. Louis, Missouri, USA. 7 Laboratory for Transcriptional Regulation, RIKEN Center for Integrative Medical Sciences-RIKEN Center for Allergy and Immunology, Yokohama, Kanagawa, Japan. 8 Friedrich Miescher Institute for Biomedical Research, Novartis Research Foundation, Basel, Switzerland. 9 University of Basel, Faculty of Sciences, Basel, Switzerland. 10 Present address: University of Edinburgh, Wellcome Trust Centre for Cell Biology, Edinburgh, UK. 11 These authors contributed equally to this work. 12 These authors jointly directed this work. Correspondence should be addressed to W.E. (wilfried.ellmeier@meduniwien.ac.at) or C.S. (christian.seiser@meduniwien.ac.at).
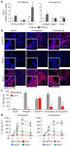
2 Supplementary Information, Boucheron et al., 2 Supplementary Figure 1 Characterization of T cells in HDAC2 cko mice. (a) Flow cytometry analysis of B220, TCRβ, CD4 and CD8α expression on splenocytes isolated from WT and HDAC2 cko mice. Flow cytometry analysis of CD4 and CD8α expression was performed on TCRβ + splenocytes. (b) Flow cytometry analysis of CD4, CD8α, CD24 and TCRβ expression on WT and HDAC2 cko thymocytes. The CD4 and CD8α plot to the right is gated on HSA lo TCRβ hi cells. (c) Flow cytometry showing intracellular HDAC2 expression levels in WT and HDAC2 cko mice DN, DP, CD4SP and CD8SP thymocyte subsets and in splenic and CD8 + T cells. CD4SP and CD8SP cells were gated on TCRβ hi subsets. The dotted vertical line separates HDAC2-negative and HDAC2-positive cells. (d) Flow cytometry analysis of CD44 and CD62L expression on TCRβ + splenic WT and HDAC2 cko and CD8 + T cells. (e) Flow cytometry analysis of CD25 and intracellular FoxP3 expression in splenic WT and HDAC2 cko T cells. (f) Flow cytometry analysis of intracellular HDAC1 and HDAC2 expression in splenic WT and HDAC2 cko and CD8 + T cells. (a,b,d-f) Numbers indicate the percentage of cells in the respective quadrants and regions. Data are representative of six (WT) and seven (HDAC2 cko mice) mice (a,b,d), of three mice (c) and of two mice (e,f) analyzed in four (a,b,d) and in one (c,e,f) experiments.
3 Supplementary Information, Boucheron et al., 3 Supplementary Figure 2 Characterization of T cell subsets in HDAC1-2 cko mice. (a) Flow cytometry analysis of TCRβ expression on WT and CD8 + T cells and on HDAC1-2 cko, CD8 + and CD8 + T cells. (b) Mean fluorescence intensity (MFI) of CD4 expression on WT T cells and on HDAC1-2 cko and CD8 + T cells. Per experimental staining group, wild-type MFI CD4 was set as 1 and the relative expression HDAC1-HDAC2-deficient and CD8 + T cells was calculated. (c) Flow cytometry analysis of CD25 and intracellular FoxP3 expression in T cells from WT T cells and in HDAC1-2 cko and CD8 + T cells (d) Percentages of CD44 hi splenic WT and CD8 + T cells and of HDAC1-2 cko, CD8 + and CD8 + T cells. (e) Flow cytometry analysis of intracellular IFN-γ and TNF-α expression in splenic WT and HDAC1-2 cko and CD8 + T cells and in HDAC1-2 cko CD8 + T cells activated ex vivo with PMA/ionomycin for 5 hours. (f) Flow cytometric bead assays showing IFN-γ, IL-4, TNFα and GM-CSF amount in the supernatant of WT and HDAC1-2 cko and CD8 + T cells and HDAC1-2 cko CD8 + T cells activated with anti-cd3 and anti-cd28 for 24 hours. (g) Flow cytometry analysis showing CD62L, CD122, CD127, CCR7 and CD27 expression on naïve (CD44 lo ) and effector/memory (CD44 hi ) splenic WT and HDAC1-2 cko and CD8 + T cells. The expression of these markers on total HDAC1-2 cko CD8 + splenocytes is shown as dashed line on top of each panel. (c,e,g) Numbers indicate the percentage of cells in the respective quadrant or regions. Data are representative (a,c,e,g) or show summary (b,d,f) of six mice (a), of seven mice (b), of two mice (c), of seven (WT) and eight (HDAC1-2 cko) mice (d), of four mice (e), of two independent samples (f) and of six mice except for CD27 (two mice) (g) that were analyzed in three (a,b), in one (c) in four (d) and in two (e,f,g) independent experiments. (b) *P < 0.05 (Wilcoxon signed rank test). Mean ± SD is shown. (d) *P < 0.05 (unpaired two-tailed Mann-Whitney test). NS, not significant.
4 Supplementary Information, Boucheron et al., 4 Supplementary Figure 3 T cell defects in HDAC1-2 cko T cells are cell-intrinsic and CD8 expression in HDAC1-2 cko T cells is dependent on Cd8 enhancer E8 I. (a) Flow cytometry analysis of B220, TCRβ, CD4 and CD8α on splenocytes isolated from CD chimeric mice that received either WT or HDAC1-2 cko CD BM cells that had been mixed at a 1:1 ratio with wild-type (WT, CD ) BM cells. Cells were gated on CD and CD subsets. Flow cytometry analysis of CD4 and CD8α expression was performed on TCRβ + splenocytes. (b) Flow cytometry analysis of CD154 and CD69 expression on WT and CD8 + T cells and on HDAC1-2 cko, CD8 + and CD8 + T cells activated with anti-cd3 and anti-cd28 for 12 hours. (c) PCR analysis of DNA isolated from WT and CD8 + T cells and B220 + B cells, and from HDAC1-2 cko, CD8 + and CD8 + T cells and B220 + B cells to detect deletion of Hdac1 and Hdac2. The lower panel shows a PCR analysis of DNA isolated from WT B cells mixed with either HDAC1 cko or HDAC2 cko T cells at the indicated ratio to determine the sensitivity of the PCR assay. The size of the PCR fragments are for Hdac1: floxed (F): 567 bp; deleted (Δ): 535 bp; for Hdac2: floxed (F): 850bp; deleted (Δ): 670bp. (d) Flow cytometry analysis of CD4 and CD8α expression on E8 I +/+ WT and E8 I / HDAC1-2 cko T cells activated with anti-cd3 and anti-cd28 for 48 hours. (a,d) Numbers in the plots indicate the percentage of cells in the respective quadrants and regions. (a-d) Data are representative of three (a) and two (b,d) mice per group analyzed in two independent experiments (a-d).
5 Supplementary Information, Boucheron et al., 5 Supplementary Figure 4 Gene expression patterns in CD4 lineage T cells in the absence of HDAC1 and HDAC2. (a) Gene expression profiles from WT T cells and from HDAC1-2 cko and CD8 + T cells were determined using Agilent arrays. Data were analyzed using Genespring software. Scatter plots show HDAC1-2 cko (Y-axis) versus WT (X-axis) T cells. The scale on the X- and Y-axis indicate expression levels (log2). Numbers at the upper-right or lower-left corners show the number of genes up- or down-regulated (fold-difference 2, P 0.05) in the absence of HDAC1 and HDAC2, respectively. (b) qrtpcr analysis showing Cd4, Cd8a and Cd8b1 expression (relative to Hprt1) in WT and CD8 + T cells and in HDAC1-2 cko, CD8 + and CD8 + T cells. Arbitrary units are shown. RNA samples were identical to the ones used for microarrays in (a). The summary of 3 independent experiments is shown. *P < 0.05, **P < 0.01 and ***P < (unpaired two-tailed Student s t-test). Mean with SD is shown. (c) Agilent microarray expression data of selected genes in WT T cells and HDAC1-2 cko and CD8 + T cells are shown. The expression levels in WT T cells were set as 1 and the relative expression in the other subsets was calculated. The numbers indicate the relative expression value. Only one representative Agilent array probe per gene is shown. The following probes are shown: Runx3 (A_55_P ), Eomes (A_55_P ), Prf1 (A_55_P ), Zbtb7b (A_55_P ), Cd4 (A_55_P ), Cd8a A_52_P443334), Cd8b1 (A_55_P ). Data show the summary of three arrays per cell population. *P < 0.05 (unpaired two-tailed Student s t-test). (d) qrtpcr analysis showing Eomes, Runx3, Tbx21 and Zbtb7b expression in naïve WT T cells and in naive HDAC1-2 cko and CD8 + ) T cells. Data show summary of 3 (for T cells) and 2 (for HDAC1-2 cko CD8 + T cells) independent biological replicas performed in two ( T cells) and one ( CD8 + T cells) experiment(s).
6 Supplementary Information, Boucheron et al., 6 Supplementary Figure 5 HDAC1-HDAC2 binding to CD8 lineage genes, the effect of GATA-3 on MS-275-induced CD8 up-regulation and a proteomic versus transcriptomic comparison. (a) qrtpcr with primers specific for Cd8a, Cd8b1, Runx3 and Eomes promoter regions and for Cd8 enhancer E8 I from chromatin of non-activated (N) and activated (A; anti-cd3 with anti-cd28 for 60 hours) wild-type (left panel) and CD8 + (right panel) T cells immunoprecipitated with anti- HDAC1 or anti-hdac2 antibodies (or IgG and IgM as control). Mean of 2 independent experiments is shown. Dots display the results of the individual experiments. One representative result of 2 independent experiments is shown for E8 I in T cells. Values are shown as % input. (b) Flow cytometry analysis showing surface CD8α expression on GFP + wild-type T cells 48 hours after transduction with either "empty" MIGR-control or with a Gata3-containing retroviral vector. MS-275 (or DMSO as carrier control) was added 24 hours after transduction. Data are representative of 4 independent experiments. The summary of all experiments is shown at the bottom. For each experiment, the percentage of CD8 + MS275-treated MIGR-transduced GFP + cells was set to 1 and the relative value of the percentage of CD8 + MS275-treated GATA3- transduced cells was calculated. *P<0.05 (one sample two-tailed t-test). Mean ± SD is shown. (c) Correlation plot of the log2-ratios for microarray transcriptomic (WT/HDAC1-2 cko) and proteomic data (WT/HDAC1-2 cko). The pearson's correlation coefficient is , and spearman's rank coefficient is
7 Supplementary Information, Boucheron et al., 7 Supplementary Table 1 Up-regulated genes 1 A_52_P Glis2 2 A_55_P Ptpla 3 A_52_P Arhgef40 4 A_51_P F13a1 5 A_55_P Plxdc2 6 A_55_P Slc6a8 7 A_51_P Cxxc5 8 A_55_P Gprasp2 9 A_55_P Trnp1 10 A_52_P H17Rik 11 A_55_P Ctnnbip1 12 A_55_P Pvrl2 13 A_51_P Vangl2 14 A_55_P Zcchc12 15 A_52_P Cd24a 16 A_55_P Large 17 A_52_P Khdc1a 18 A_52_P Tcf7l1 19 A_51_P Cap2 20 A_55_P Eno2 21 A_51_P Grtp1 22 A_55_P N06Rik 23 A_55_P Mn1 24 A_55_P Trip10 25 A_51_P Cebpd 26 A_55_P Dtl 27 A_51_P Sept4 28 A_51_P Ccdc102a 29 A_55_P Slc6a19 30 A_55_P Nlgn2 31 A_52_P Mex3a 32 A_51_P Ddah2 33 A_55_P Bmf 34 A_51_P Rundc3a 35 A_51_P N04Rik 36 A_55_P Tbkbp1 37 A_52_P Nedd4 38 A_55_P R A_55_P Tmem151b 40 A_55_P Emid1 41 A_55_P Zfp A_51_P Smo 43 A_51_P Trpm1 44 A_55_P Tmed1 45 A_51_P Pipox 46 A_55_P Rsph1 47 A_55_P Acvr2b 48 A_51_P Syngr1 49 A_51_P Wdr35 50 A_55_P Cxadr 51 A_55_P Ier5l 52 A_66_P Kit 53 A_55_P Fbxo44 54 A_51_P Marcksl1 55 A_51_P Slc35f5 56 A_51_P Slc6a13 57 A_55_P Pnck 58 A_55_P Aldh7a1 59 A_55_P Pacsin3 60 A_52_P Vat1 61 A_51_P Spag5 62 A_51_P Padi3 63 A_51_P Dntt 64 A_55_P Pkdcc 65 A_55_P Ptprf 66 A_52_P Wipi1 67 A_51_P Sema4g 68 A_55_P Zcchc24 69 A_55_P Ddc 70 A_55_P Gpr83 71 A_55_P Adssl1 72 A_51_P Arhgef25 73 A_55_P Pmepa1 74 A_51_P Igsf3 75 A_55_P Agrn 76 A_52_P Klhdc8b 77 A_55_P Tmem53 78 A_55_P Itih5 79 A_55_P Eng 80 A_55_P Kank2 81 A_51_P Whrn 82 A_55_P Rcor2 83 A_52_P Lmna 84 A_66_P Ogfrl1 85 A_55_P AW A_52_P Rbm24 87 A_51_P Rasgrp4 88 A_55_P Egfl7
8 Supplementary Information, Boucheron et al., 8 89 A_55_P L24Rik 90 A_55_P Pdzd4 91 A_51_P Gadd45g 92 A_51_P Cldn10 93 A_55_P C A_55_P Ptprs 95 A_51_P Ikbip 96 A_55_P Cdk1 97 A_51_P Klc3 98 A_55_P Gtf2ird1 99 A_52_P Hist1h2ab 100 A_55_P G10Rik 101 A_52_P Padi4 102 A_55_P Pkig 103 A_55_P Reep1 104 A_51_P Chst A_55_P Zfp A_52_P Ccdc A_52_P Ska1 108 A_51_P Zfp A_51_P Rilpl1 110 A_66_P Cdc6 111 A_55_P Trp53inp2 112 A_51_P H2-Oa 113 A_52_P Chl1 114 A_55_P Rcn3 115 A_51_P Pask 116 A_55_P Gpr A_51_P Tubb6 118 A_55_P Dbn1 119 A_55_P B930036G03Rik 120 A_55_P Lrfn4 121 A_66_P Tlr A_51_P Depdc1b 123 A_52_P Khdc1c 124 A_55_P Plxnb2 125 A_51_P Usp A_51_P As3mt 127 A_51_P St3gal5 128 A_55_P Selenbp1 129 A_51_P Neurl2 130 A_55_P Aldh3b1 131 A_51_P Pygl 132 A_51_P Slc7a7 133 A_55_P Tex9 134 A_51_P Cul7 135 A_55_P Slc22a A_51_P Plekho1 137 A_55_P O12Rik 138 A_51_P Lilrb4 139 A_51_P Hist1h1a 140 A_55_P Dab2ip 141 A_51_P Bub1 142 A_55_P Plekhh3 143 A_52_P Ccnjl 144 A_51_P Slc25a A_55_P Plekhg5 146 A_51_P Phlda3 147 A_55_P Rad51ap1 148 A_51_P Nusap1 149 A_52_P Art1 150 A_55_P Dnahc A_55_P Cd8b1 152 A_51_P Cacnb3 153 A_52_P Dyrk3 154 A_55_P Egln3 155 A_55_P Crispld2 156 A_52_P Neil3 157 A_55_P Tmem176b 158 A_66_P O10Rik 159 A_55_P Tcn2 160 A_55_P Sox4 161 A_51_P Lrp A_51_P Acadl 163 A_55_P Rbpms2 164 A_51_P Cd A_51_P Amigo2 166 A_52_P Ift A_52_P Slc15a1 168 A_51_P Nr2f6 169 A_55_P Sh3gl3 170 A_66_P B930049P21Rik 171 A_51_P Tspan6 172 A_55_P Zfp A_55_P St6galnac6 174 A_55_P Ccl27a 175 A_51_P Ptgr1 176 A_55_P Mapre3 177 A_55_P Ephb6 178 A_52_P Hist1h2ak 179 A_55_P Ikbip 180 A_55_P Gm5771
9 Supplementary Information, Boucheron et al., A_55_P Uhrf1 182 A_51_P Kif A_55_P Pafah1b3 184 A_55_P Gm A_55_P Rrm2 186 A_51_P Purg 187 A_55_P Rpgr 188 A_55_P Pter 189 A_52_P Aph1b 190 A_55_P Adam A_51_P Tubb2b 192 A_55_P Hist1h3i 193 A_51_P Cenph 194 A_51_P Bend5 195 A_52_P Luzp1 196 A_51_P Kif7 197 A_55_P Ssh3 198 A_55_P Pdlim7 199 A_52_P Akap2 200 A_51_P Ppm1n 201 A_51_P Nphp3 202 A_51_P Rad A_51_P Ctsl 204 A_55_P Arhgef A_51_P Agpat4 206 A_55_P Tspan3 207 A_55_P Tox2 208 A_55_P Gfi1 209 A_55_P Spsb2 210 A_55_P B20Rik 211 A_51_P Mns1 212 A_55_P Ntn1 213 A_55_P Hist1h2ag 214 A_51_P Cks1b 215 A_52_P Atat1 216 A_55_P Txnrd3 217 A_51_P Hk2 218 A_55_P E21Rik 219 A_52_P E07Rik 220 A_55_P Abcb A_51_P Litaf 222 A_55_P Hist1h2an 223 A_55_P Wdr A_55_P Tns1 225 A_52_P Rad54b 226 A_52_P Gm A_52_P Intu 228 A_51_P Gpr A_52_P Vegfb 230 A_52_P Zdhhc1 231 A_52_P Celsr2 232 A_51_P Morn1 233 A_52_P Klf A_55_P Armcx6 235 A_51_P Ift A_55_P Ggta1 237 A_51_P Dos 238 A_52_P Abca1 239 A_55_P Ift A_55_P Colq 241 A_51_P Plscr3 242 A_51_P Ehd1 243 A_55_P Ncaph 244 A_51_P Ttc8 245 A_51_P Stard A_51_P Cdca5 247 A_52_P Suv39h2 248 A_51_P Nrn1 249 A_55_P Gm A_55_P E13Rik 251 A_55_P Cnn3 252 A_55_P Prx 253 A_51_P Ggt1 254 A_55_P Raph1 255 A_52_P Sgol2 256 A_55_P Epb4,1l1 257 A_55_P G19Rik 258 A_55_P Myo18a 259 A_51_P Shank3 260 A_55_P Dapk1 261 A_55_P Fam131a 262 A_55_P Tmem A_51_P Cox6b2 264 A_55_P Slc12a4 265 A_55_P Lrrc4b 266 A_55_P Usp2 267 A_51_P Mxd1 268 A_51_P Fbxo A_51_P E130303B06Rik 270 A_51_P Ift A_55_P Hist2h3c1 272 A_55_P Dtx3
10 Supplementary Information, Boucheron et al., A_55_P Tspan A_51_P Slc4a8 275 A_55_P K19Rik 276 A_55_P Hist1h2bg 277 A_51_P Elovl6 278 A_66_P Numbl 279 A_52_P Sdc3 280 A_55_P Rufy3 281 A_52_P Clip2 282 A_55_P Tacc2 283 A_55_P Leprel4 284 A_55_P Gm A_52_P Cdc6 286 A_55_P H2-Ob 287 A_55_P Azi1 288 A_55_P Gprasp1 289 A_55_P Ankrd13d 290 A_51_P Kntc1 291 A_51_P Usp6nl 292 A_55_P Wdr A_51_P Srsf A_55_P Gata1 295 A_55_P Greb1 296 A_55_P Pdlim7 297 A_55_P Rgs A_55_P A07Rik 299 A_55_P Pfn2 300 A_55_P Hpse 301 A_55_P Ppp1r16a 302 A_51_P Prelid2 303 A_55_P Prkar2b 304 A_55_P Cul9 305 A_55_P Hist1h2ah 306 A_55_P Reck 307 A_51_P Cenpp 308 A_55_P Agl 309 A_55_P Maml3 310 A_51_P Prpf40b 311 A_51_P Scamp1 312 A_55_P Tdrd5 313 A_55_P H08Rik 314 A_52_P Fanca 315 A_66_P Arl3 316 A_52_P Timp2 317 A_52_P G0s2 318 A_51_P Gins1 319 A_55_P Myb 320 A_55_P Pcyox1 321 A_55_P Ccdc A_66_P Map3k8 323 A_51_P Chchd A_55_P Hdac6 325 A_51_P BC A_55_P Smox 327 A_52_P Ckb 328 A_51_P Tm6sf1 329 A_55_P Dip2a 330 A_51_P Chtf A_51_P Cand2 332 A_51_P Odf3b 333 A_55_P St3gal3 334 A_51_P D01Rik 335 A_55_P Zfp A_51_P Abi2 337 A_55_P Alms1 338 A_55_P Ift A_55_P AI A_51_P Trip A_55_P Ccdc A_51_P Fam195a 343 A_55_P Gsn 344 A_51_P Zfp A_55_P Gmnn 346 A_66_P Hist2h2ac 347 A_51_P N4bp3 348 A_51_P Zfyve A_55_P Hist1h2af 350 A_55_P Gatsl2 351 A_55_P Aqp A_51_P Tst 353 A_55_P Tox 354 A_51_P Scd2 355 A_51_P Akr1c A_51_P Ndrg1 357 A_51_P BC A_55_P H2afv 359 A_55_P Garnl3 360 A_55_P Aplp1 361 A_55_P Sgsm2 362 A_51_P Cdca8 363 A_55_P Gale 364 A_55_P Arvcf
11 Supplementary Information, Boucheron et al., A_55_P Fbxl A_51_P H22Rik 367 A_55_P Caskin1 368 A_51_P E2f3 369 A_55_P Hist1h3f 370 A_51_P Wls 371 A_55_P Gm A_51_P Plk1 373 A_55_P Atp1a3 374 A_51_P Nr4a1 375 A_55_P Nelf 376 A_51_P Itm2a 377 A_55_P Camkk1 378 A_52_P Akap17b 379 A_55_P Mst1 380 A_51_P Fhl2 381 A_55_P Gstm1 382 A_55_P Trp53i A_51_P Tmem9 384 A_55_P Dlg3 385 A_55_P Ahdc1 386 A_51_P Ssx2ip 387 A_52_P Zscan A_66_P Ttc3 389 A_55_P Gas8 390 A_51_P Zfp518b 391 A_51_P Trib1 392 A_51_P Acot7 393 A_51_P Atpif1 394 A_55_P Rps6ka5 395 A_55_P Rims3 396 A_52_P D08Rik 397 A_51_P Dnalc4 398 A_55_P C02Rik 399 A_55_P Lactb 400 A_52_P H2afj 401 A_51_P Cuedc2 402 A_51_P Tlr2 403 A_55_P Tjp3 404 A_51_P E130308A19Rik 405 A_55_P Slc16a3 406 A_55_P Hist2h2aa2 407 A_55_P Paox 408 A_51_P Katnal2 409 A_55_P Hist1h4k 410 A_55_P Cerk 411 A_51_P Gm A_52_P Hic2 413 A_55_P Chka 414 A_52_P Atp5s 415 A_66_P Crxos1 416 A_51_P Ern1 417 A_51_P Chaf1b 418 A_55_P Lman2l 419 A_55_P Sirt3 420 A_55_P Zfpm1 421 A_55_P Dmwd 422 A_55_P J21Rik 423 A_51_P Ncapd2 424 A_51_P Gabbr1 425 A_55_P Pbx3 426 A_51_P Grik5 427 A_55_P Mark4 428 A_51_P Socs3 429 A_55_P Rarg 430 A_51_P Clcn2 431 A_55_P Renbp 432 A_66_P Dhfr 433 A_55_P Casc1 434 A_51_P Pcbp4 435 A_55_P Shf 436 A_55_P Tmem39b 437 A_55_P Mgmt 438 A_55_P Hmgn2 439 A_55_P A530028O A_55_P Lzts2 441 A_51_P Skap2 442 A_55_P Ston1 443 A_55_P Hmgb3 444 A_55_P E2f1 445 A_55_P S100a A_52_P Dpcd 447 A_55_P Slc30a4 448 A_66_P I02Rik 449 A_55_P Hist1h4j 450 A_55_P Bai2
12 Supplementary Information, Boucheron et al., 12 Supplementary Table 1 (continued) Down-regulated gene 1 A_55_P Cyp2s1 2 A_51_P Bcap29 3 A_55_P Slc28a2 4 A_51_P B13Rik 5 A_52_P Tnfrsf26 6 A_52_P Zfp287 7 A_55_P C18Rik 8 A_52_P Rragb 9 A_55_P K14Rik 10 A_51_P Fam118a 11 A_52_P G20Rik 12 A_51_P Zfp A_66_P Inadl 14 A_55_P Tsen54 15 A_51_P Slc19a1 16 A_51_P Snx15 17 A_51_P Slc46a3 18 A_52_P Map3k5 19 A_52_P Ccdc9 20 A_52_P C19Rik 21 A_66_P B3galtl 22 A_55_P Btbd19 23 A_51_P Tmem42 24 A_55_P Cryl1 25 A_51_P Npc2 26 A_51_P Dstn 27 A_51_P Thnsl1 28 A_51_P Zscan12 29 A_51_P Ubxn8 30 A_51_P Lhpp 31 A_51_P Trim68 32 A_51_P Pmm2 33 A_55_P Rapgef6 34 A_51_P BC A_51_P Dnajb13 36 A_55_P Cr1l 37 A_55_P Ctss 38 A_55_P LOC A_51_P Vkorc1 40 A_55_P Rpl22l1 41 A_55_P Arhgap29 42 A_51_P Slc25a45 43 A_52_P Rrp12 44 A_51_P Anapc11 45 A_55_P LOC A_55_P Il6ra 47 A_55_P Zbtb16 48 A_55_P Gm A_51_P Dennd2d 50 A_51_P Praf2 51 A_51_P Rnf144a 52 A_55_P Pde3b 53 A_51_P Neurl3 54 A_52_P Rundc3b 55 A_55_P Etv3 56 A_52_P Evi2b 57 A_55_P G13Rik 58 A_55_P I08Rik 59 A_52_P Trat1 60 A_66_P D01Rik 61 A_51_P Klhl6 62 A_52_P Tmem8 63 A_65_P Scrn3 64 A_51_P B3galt4 65 A_51_P F06Rik 66 A_51_P Abcb4 67 A_55_P Nek3 68 A_51_P Mpzl3 69 A_55_P Ibtk 70 A_51_P Amz2 71 A_51_P Filip1l 72 A_55_P Hcst 73 A_55_P Tmub1 74 A_55_P Hdac1 75 A_51_P Sdcbp2 76 A_55_P Vipr1 77 A_55_P Pcdhgb1 78 A_51_P F03Rik 79 A_55_P Hif3a 80 A_51_P Hsd11b1 81 A_51_P S1pr4 82 A_55_P Sntb1 83 A_55_P Tmem20 84 A_55_P Lrrc61 85 A_51_P Cdc42ep3 86 A_51_P Smyd2 87 A_51_P Rpp38 88 A_52_P Zc3h12d
13 Supplementary Information, Boucheron et al., A_51_P Daglb 90 A_55_P Trib3 91 A_51_P Mmaa 92 A_51_P Gbp2 93 A_51_P Ppic 94 A_51_P Afp 95 A_51_P Abcb9 96 A_52_P Ydjc 97 A_55_P Gpr137b-ps 98 A_55_P Olfr A_66_P Atp8b4 100 A_52_P Gm A_55_P Fam89a 102 A_52_P Hsd17b A_51_P Hsdl1 104 A_55_P Unc5a 105 A_55_P M22Rik 106 A_52_P Slc35d2 107 A_51_P Abhd A_51_P Ank 109 A_55_P Art4 110 A_51_P Pecr 111 A_51_P Sidt1 112 A_55_P Nckap1 113 A_55_P Slc16a A_51_P St8sia6 115 A_55_P Atg A_66_P Ntrk3 117 A_52_P Zfp3 118 A_51_P P10Rik 119 A_51_P Ctnna1 120 A_51_P Nipa1 121 A_52_P Amica1 122 A_55_P Apol7b 123 A_51_P Golm1 124 A_55_P L09Rik 125 A_55_P Dhrs A_55_P Abcg2 127 A_51_P Fam46a 128 A_51_P Mansc1 129 A_55_P Cd A_55_P G24Rik 131 A_55_P A130040M12Rik 132 A_55_P B230216N24Rik 133 A_52_P Wscd2 134 A_55_P Gpr137b 135 A_52_P Gpr A_51_P Abcb1b 137 A_55_P AI A_66_P Daf2 139 A_55_P Grm6 140 A_51_P Appl2 141 A_51_P Galnt9 142 A_66_P Kctd A_51_P Mgst3 144 A_51_P Tbc1d A_51_P Rgmb 146 A_55_P Prg4 147 A_55_P F24Rik 148 A_51_P Amz1 149 A_55_P Fasl 150 A_51_P Nme4 151 A_55_P App 152 A_51_P Rnf A_55_P Bend6 154 A_52_P Adh1 155 A_55_P Gm A_55_P Gm A_55_P Ntrk3 158 A_52_P Gm A_55_P Kcnj A_52_P Sh3bgrl2 161 A_51_P Prss A_55_P Chi3l7 Supplementary Table 1 lists all genes that are differentially expressed (>2 Fold change, P<0.05) between HDAC1-2 cko (cko) T cells and WT T cells. Positive and negative values indicate up- and down-regulation, respectively, in the absence of HDAC1/2. Gene expression profiles were determined using Agilent arrays and GeneSpring software. Only those probes with assigned gene names are listed (long intergenic non-coding (linc) RNAs are excluded from the list).
14 Supplementary Information, Boucheron et al., 14 Supplementary Table 2 Up-regulated proteins 1 Q8BH Rcn3 2 P Ctsg 3 P Ctse 4 Q9DBX Susd2 5 P Hbb-b1 6 P , P Igh-6 7 Q9Z Pf4 8 Q91WU As3mt 9 Q9WUU Ctsz 10 P Grn 11 Q Prtn3 12 Q8K023-[1,2] 2.24 Akr1c18 13 Q9D0J Ptms 14 P Igj 15 P Hba 16 P Lgals3 17 P P Ctsh 19 Q8VCM Fgg 20 Q9DBF1-[1,2] 1.98 Aldh7a1 21 Q8K0E Fgb 22 P31996-[1,2] 1.91 Cd68 23 Q9D394-[1,2,3,4] 1.91 Rufy3 24 Q9BDB7-[1,2] 1.90 IFI44L 25 Q8K558-[1,2] 1.89 Treml1 26 P Marcksl1 27 Q Isg15 28 P Glb1 29 Q Sord 30 Q99L PAGR1 31 P Fgr 32 Q8BG07-[1,2] 1.80 Pld4 33 P0CW Ly6c2 34 Q Acp5 35 O Chi3l3 36 P Stom 37 P17710-[1,2,4] 1.76 Hk1 38 P17047-[1,2,3] 1.75 Lamp2 39 P Tgm2 40 P Aldh2 41 P Ctsb 42 O Myadm 43 Q9DAW Cnn3 44 P Kif4 45 P14434, P H2-Aa 46 P Etv6 47 Q Hp 48 Q9D8I Pacap 49 Q Psap 50 P Mmp9 51 Q Ndrg1 52 P S100a4 53 P S100a11 54 Q Vat1 55 Q3TRM Hk3 56 Q8VCI Plbd1 57 Q Ckb 58 Q Hist1h1t 59 Q91W Klc3 60 Q9WUL Arl3 61 P Acp2 62 Q91XV Basp1 63 Q9QUM Itga2b 64 P Fkbp2 65 Q Pafah1b3 66 P Gsn 67 Q8K4X Agpat4 68 Q8R2U Nudt4 69 O Ppt1 70 Q8BTV Tusc3 71 Q8BV Ifi44 72 P Thbs1 73 P Ctsd 74 O Dnmt3a 75 Q8VC Akr1c13 76 Q Satb1 77 P28650-[1,2] 1.56 Adssl1 78 P Lmna 79 O Atpif1 80 Q9WVK Ehd1 81 Q8R Sccpdh 82 Q9ET Pygl 83 Q Vav2 84 Q99LD Ddah2 85 Q8VEB Pla2g15 86 P Gstm1 87 Q99PL Rrbp1 88 Q9JJV2-[1,2,3] 1.52 Pfn2
15 Supplementary Information, Boucheron et al., Q9CQF Pcyox1 90 O Pld3 91 P Nedd4 92 Q9WV Asah1 93 Q9Z0H8-[1,2] 1.49 Clip2 94 P Ctsa 95 Q9QYG0-[1,2] 1.48 Ndrg2 96 Q6DID7-[1,2] 1.48 Wls 97 Q6ZQ Cand2 98 Q920A Scpep1 99 P Ca2 100 Q4FZC Syne3 101 Q921J Ube2s 102 Q8R Gale 103 P Casp3 104 P Stmn1 105 P97377-[1,2] 1.46 Cdk2 106 O Hnrnph1 107 Q9CQM Txndc Q7TMR Prcp 109 Q Vcl 110 P Hsp90b1 111 O Itgb3 112 Q9JJC Rilpl1 113 Q91W Txndc5 114 Q60848-[1,2] 1.44 Hells 115 O Il4i1 116 Q3TB Plekhf1 117 Q9ER Tor3a 118 Q8BGA Q Ggt1 120 Q Rcn1 121 Q9D7V Naaa 122 Q Ifit1 123 Q8BY Hat1 124 Q3U5Q Cmpk2 125 P Gaa 126 O Vamp4 127 Q8R2P Klhl P Sts 129 Q7TMM Tubb2a 130 Q6DFV5-[1,2,3] 1.40 Helz 131 P13597-[1,2] 1.39 Icam1 132 Q9ER Rtp4 133 P Cdk1 134 Q4QQM Trp53i Q8VDF Uhrf1 136 Q9CYL Glipr2 137 P Cd Q9WVJ3-[1,2] 1.38 Cpq 139 Q Top2a 140 Q9CPY Mrpl Q8C1B7-[1,2,3] 1.37 Sept Q8CAB Gatsl2 143 Q Sema4a 144 O Ppt2 145 Q91WC Setd3 146 Q2HXL Edem3 147 Q9ET Dpp7 148 P Tyms 149 Q3UND0-[1,2] 1.36 Skap2 150 P Urod 151 Q9R1Q Plp2 152 O Atox1 153 Q9DC Ergic1 154 P Atp8a1 155 Q9DBY Cbx6 156 Q91V Acot7 157 Q99KC Vwa5a 158 Q9WVQ Apip 159 P97822-[1,2] 1.34 Anp32e 160 Q8R3L2-[1,2,3,4,5] 1.34 Tcf O Man2b1 162 P Ada 163 P Map4 164 Q9QZ Iigp1 165 P Tubb5 166 Q ,34 Zyx 167 E9Q355-[1.2,3] 1.33 Catsperg1 168 P P4hb 169 Q9WVS8-[1,4,5] 1.33 Mapk7 170 Q Lgals3bp 171 O Man2b2 172 P Aco1 173 Q Acads 174 P0C7N Psmg4 175 P Mapkapk2 176 Q8K3C Rnasek 177 P Pfkm 178 P Atp6v0d1 179 Q9D3G Stx Q8BP27-[1,2] 1.33 Sfr1
16 Supplementary Information, Boucheron et al., Q922R Pdia6 182 Q8C0L Paox 183 P Mapk O Ugcg 185 P Fas 186 Q Serpinb6 187 P Pdia4 188 O Ikzf3 189 Q9Z0M Lipa 190 Q9QXT Cnpy2 191 Q8BIW Prune 192 P Hsd17b4 193 O Npc1 194 P Gusb 195 Q9JKR Hyou1 196 Q9JJZ Ube2j1 197 Q Ppp1r14b 198 Q8K2Q Brox 199 Q9CQU Zwint 200 A2AJI0-[1,2] 1.31 Map7d1 201 P Acadl 202 Q Cdk6 203 Q6ZQK Ncapd3 204 O08795-[1,2] 1.30 Prkcsh 205 Q6PAL Ahdc1 206 Q99L Hibadh Supplementary Table 2 (continued) Down-regulated proteins 1 Q9CY66-[1,2,3] Gar1 2 Q8BGX Q9D1H Ndufaf4 4 Q3TC Ccdc127 5 Q6AXF Sidt1 6 P Evl 7 Q5M8N Sdr39u1 8 Q9CQS Nop10 9 Q9DCT6-[1,2] Bap18 10 Q8BMS Rassf2 11 Q9ESX Dkc1 12 Q9CRB Nhp2 13 Q9Z2I Suclg2 14 P Apex1 15 Q9Z0E Gbp2 16 Q8VE Q8BR Suds3 18 Q8BFP Pdk1 19 Q8BTX Hsdl1 20 P Timm13 21 Q9CZL Pcbd2 22 Q9Z2L Crlf3 23 Q9D Dapl1 24 P Eci1 25 P Cd28 26 Q9D Mthfs 27 O Hdac1 28 Q91VV4-[1,2] Dennd2 29 Q Pdcd4 30 Q923B Ggact 31 P Zfp58 32 Q99KE Me2 33 Q91WE Snx15 34 Q9QYR Acot2 35 Q9EQ Hsd17b11 36 Q8K Acad10 37 Q924K Mta3 38 Q5DU31-[1,2] Ipcef1 39 Q9Z2D8-[1,2] Mbd3 40 Q8K2T P Prkacb 42 P Hdac2 43 Q9WV Nme4 44 Q9D7S7-[1,2] Rpl22l1 45 P Ifrd1 46 Q8C5P Q8R4N Clybl 48 P Adh1 49 P15092-[1,2] Ifi204 Supplementary Table 2 lists the proteins that are differentially expression (>1.3 Fold change, FC; P<0.05) between HDAC1-1 cko (cko) T cells and WT T cells.
17 Supplementary Information, Boucheron et al., 17 Supplementary Table 3 List of up-regulated proteins with normal RNA expression 1 P Ctsg 2 P Ctse 3 Q9DBX Susd2 4 Q9Z Pf4 5 Q9WUU Ctsz 6 P Grn 7 Q Prtn3 8 Q8K023-1,Q8K Akr1c18 9 Q9D0J Fut4 10 P Igj 11 P P Ctsh 13 Q8VCM Gng8 14 Q8K0E Fgb 15 P ,P Cd68 16 Q8K558-1,Q8K Treml1 17 Q Isg15 18 P Glb1 19 Q Dmd 20 Q99L E17 Rik 21 P Fgr 22 Q8BG07-1,Q8BG Pld4 23 P0CW Ly6c1 24 Q Acp5 25 O Chi3l3 26 P Stom 27 P ,P ,P Hk1 28 P ,P ,P Lamp2 29 P Tgm2 30 P Aldh2 31 P Ctsb 32 O Myadm 33 P14434,P H2-Aa 34 P Chrnd 35 Q Hp 36 Q9D8I M0 9Rik 37 Q Psap 38 P Mmp9 39 P S100a4 40 Q3TRM Hk3 41 Q8VCI Plbd1 42 P Acp2 43 Q91XV Basp1 44 Q9QUM Itga2b 45 P Fkbp2 46 Q8R2U Nudt4 47 O Entpd6 48 Q8BTV Tusc3 49 Q8BV Ifi44 50 P Ctsd 51 O Dnmt3a 52 Q8VC Akr1c13 53 Q Satb1 54 Q8R Sccpdh 55 Q Vav2 56 Q8VEB Pla2g15 57 P Cmah 58 Q99PL Rrbp1 59 O Pld3 60 Q9WV Asah1 61 P Ctsa 62 Q9QYG0-1,Q9QYG Ndrg2 63 Q920A Scpep1 64 P Car2 65 Q4FZC I19 Rik 66 Q921J Ube2s 67 P Casp3 68 P Stmn1 69 P ,P Cdk2 70 O Hnrnph1 71 Q9CQM Txndc17 72 Q7TMR Prcp 73 Q Vcl 74 P Hsp90b1 75 O Itgb3 76 Q91W Txndc5 77 Q ,Q Hells 78 O Il4i1 79 Q3TB Plekhf1 80 Q9ER Tor3a E130309D0 81 Q8BGA Rik
18 Supplementary Information, Boucheron et al., Q Rcn1 83 Q9D7V Naaa 84 Q Ifit1 85 Q8BY Hat1 86 Q3U5Q Cmpk2 87 P Gaa 88 O Vamp4 89 Q8R2P Klhl25 90 Q6DFV5-1.Q6DFV5-2.Q6DFV Helz 91 P P Icam1 92 Q9ER Rtp4 93 Q9CYL Glipr2 94 P Cd38 95 Q9WVJ3-1.Q9WVJ Pgcp 96 Q Top2a 97 Q9CPY Mrpl51 98 Q8C1B7-1.Q8C1B7-2,Q8C1B Sep Q Sema4a 100 O Ppt2 101 Q91WC Setd3 102 Q2HXL Edem3 103 Q9ET Dpp7 104 P Tyms 105 P Urod 106 Q9R1Q Plp2 107 O Atox1 108 Q9DC Ergic1 109 P Atp8a1 110 Q9DBY Cbx6 111 Q99KC Vwa5a 112 Q9WVQ Apip 113 P ,P Anp32e 114 Q8R3L2-1,Q8R3L2-2,Q8R3L2-3,Q8R3L2-4,Q8R3L Tcf O Man2b1 116 P Ada 117 P Mtap4 118 Q9QZ Cyp2f2 119 P Tubb5 120 Q Zyx 121 P Dld 122 Q9WVS8-1,Q9WVS8-4,Q9WVS Mapk7 123 Q Lgals3bp 123 Q Lgals3bp 124 O Man2b2 125 P Hcn1 126 Q Acads 127 P0C7N Psmg4 128 Q8K3C Rnasek 129 P Mapkapk2 130 P Pfkm 131 P Atp6v0d1 Q8BP27-1,Q8BP E Rik 133 Q8C0L Paox 134 P Mapk O Ugcg 136 P Fas 137 Q Serpinb6a 138 P Pdia4 139 O Ikzf3 140 Q9Z0M Lipa 141 Q9QXT Cnpy2 142 Q8BIW Gli3 143 P Hsd17b4 144 O Npc1 145 P Gusb 146 Q9JKR Apoc3 147 Q9JJZ Ube2j1 148 Q Ppp1r14b 149 Q9CQU Zwint 150 A2AJI0-1,A2AJI Mtap7d1 151 Q Cdk6 152 O ,O Prkcsh 153 Q99L Hibadh
19 19 Supplementary Table 3 (continued) List of down-regulated proteins with normal mrna expression Q9CY66-1,Q9CY ,Q9CY Gar J Q8BGX Rik 156 Q9D1H Ndufaf4 157 Q3TC Ccdc P Evl 159 Q5M8N Sdr39u1 160 Q9CQS Nop Q8BMS Rassf2 162 Q9ESX Dkc1 163 Q9CRB Nhp2 164 Q9Z2I Suclg2 165 P Apex1 166 Q8VE C030006K1 1Rik 167 Q8BR Suds3 168 Q8BFP Pdk1 169 P Timm Q9CZL Pcbd2 171 Q9Z2L Hars 172 Q9D Dapl1 173 P Eci1 174 P Cd Q9D Gm Q Apoc2 177 Q923B A2ld1 178 P Zfp Q99KE Me2 180 Q9QYR Acot2 181 Q8K Acad Q924K Mta3 183 Q5DU31-1.Q5DU Ipcef1 184 Q9Z2D8-1.Q9Z2D Mbd3 185 Q8K2T AI P Prkacb 187 P Hdac2 188 P Ifrd1 189 Q8R4N Clybl Supplementary Table 3 lists the genes that are differentially expressed at the protein level (>1.3 Fold change, FC; P<0.05) but not at the mrna level (defined as >2 Fold change, P<0.05) between HDAC1-2 cko (cko) T cells and WT T cells.
20 20 Supplementary Table 4 List of antibodies used in this study Antigen/Name Clone Fluorochrome Vendor CD4 RM4-5 PE, PE-Cy7 ebioscience CD8α APC ebioscience CD25 PC PE ebioscience CD FITC ebioscience CD69 H1.2F3 FITC ebioscience Eomesodermin Dan11mag APC ebioscience FoxP3 FJK-16s APC ebioscience TNFα MP6-XT22 FITC ebioscience TCRβ H APC-e780 ebioscience TCR Vα2 B20.1 PE ebioscience CD24 M1.69 PE-Cy5 Biolegend CD APC BD Bioscience CD8α Biotin BD Bioscience CD8β H FITC BD Bioscience CD11b M1/70 Biotin BD Bioscience CD11c HL3 Biotin BD Bioscience CD16/CD32 2.4G2 - BD Bioscience CD44 IM7 V500 BD Bioscience CD45R/B220 RA3-6B2 Biotin, FITC BD Bioscience CD62L MEL-14 PE, APC BD Bioscience CD154 MR1 PE BD Bioscience GATA3 L PE BD Bioscience Gr-1 RB6-8C5 PE BD Bioscience Granzyme B GB11 V450, PE, APC BD Bioscience IFNγ XMG1.2 FITC, V450 BD Bioscience Mouse IgG1 A BD Bioscience NK1.1 PK136 Biotin BD Bioscience T-bet 4B10 V450, APC BD Bioscience TCRβ H V450 BD Bioscience Ter119 TER-119 Biotin BD Bioscience HDAC1 ABE260 - Millipore HDAC2 3F3 - Millipore Anti-rabbit IgG1 (H+L) - FITC Invitrogen
21 Supplementary Information, Boucheron et al., 21 Supplementary Table 5 List of primers used in this study Locus Forward sequence (5 3 ) Reverse sequence (5 3 ) Method Ref # Cd4 TTCCTGGCTTGCGTGCTGGG TTCTGCATCCGGTGGGGGCA qrtpcr Cd8a AGCCAGTGCTGCGAACTCCCT TCGGCTCCTGTGGTAGCAGATGA qrtpcr Cd8b1 GGGACGAAGCTGACTGTGGTTGA GGGTGGGGGAACGGGCATTG qrtpcr Eomes GCCTACCAAAACACGGATA TCTGTTGGGGTGAGAGGAG qrtpcr 1 GzmB CCACTCTCGACCCTACATGG GGCCCCCAAAGTGACATTTATT qrtpcr 2 Hprt GATACAGGCCAGACTTTGTTG GGTAGGCTGGCCTATAGGCT qrtpcr 3 Prf1 TTTCGCCTGGTACAAAAACC CGTTCAGGCAGTCTCCTACC qrtpcr 1 Runx3 GTCAGCGTGCGACATGGCTTCCAACAG AGCACGTCCATCGAGCGCACTTCGG qrtpcr 4 Tbx21 CAACAACCCCTTTGCCAAAG TCCCCCAAGCAGTTGACAGT qrtpcr 5 Zbtb7b ACCCCGACCTGATGGCTTAC GCGTTCTCCTGTGTGCTTCC qrtpcr 6 Cd8a promoter CCACTTGCTCTGCAAGGGTGCA GTGGGGGAGGCGTGTAGGGT ChIP Cd8b1 promoter TCTTCATGGGTTCTTGATGGGGCTT AAAGTGAGGCAAAGCCAAACCTGG ChIP E8 I #26 GGGGTGCCTCAACCTGCTTTGT TGTGGCGGGCTTCATGGCATAG ChIP E8 I #33 ATGGCAAAGTCTGCCGGCTTCC TGGCCTGCTCCCAGGTTCCTTTT ChIP Runx3 promoter Eomes promoter Ifng promoter Gzmb promoter TCCTGTAGCCCCACTTTC TCGTCTATTCTGCCCTCG ChIP TATTAATTGGATCGGGTCGCCTGC ACCATGTTCGCAGACTTCAAACCC ChIP 7 ACTCTAACATGCCACAAAACCATAG CTTCCAGTTTTATACCTGATCGAAG ChIP 8 TGATGACGTCTTCTGAGTACTTGTG ACTCTGATACCATAGGCTACAAACC ChIP 8 Prf1 TSS CAGGGCAGGAAGTAGTAATGATATG CTTCCTCCTCCTTACCTGAAGTC ChIP 8 Tcrb enhancer AGAATGGCCACCTGCCATAG GATGTGGCAAGTGTGGTTCCCAAA ChIP 9 Hdac1 GATGTGGCAAGTGTGGTTCCCAAA CCTGTGTCATTAGAATCTACTT Genotyping Hdac2 CCCTTTAGGTGTGAGTACAT AACCTGGAGAGGACAGCAAA Genotyping Hdac1 deletion Hdac2 deletion GGTAGTTCACAGCATAGTACTT GTTACGTCAATGACATCGTCTT Genotyping CCACAGGGAAAAGGAAACAA AACCTGGAGAGGACAGCAAA Genotyping Primer pair E8 I #26: maps in close proximity to region g of Sato et al 10. Primer pair E8 I #33: overlaps with region i of Sato et al. 4 and region h from Hassan et al 11. References for primers 1. Tayade, C. et al. Differential transcription of Eomes and T-bet during maturation of mouse uterine natural killer cells. J Leukoc Biol 78, (2005). 2. Zhu, Y. et al. T-bet and eomesodermin are required for T cell-mediated antitumor immune responses. J Immunol 185, (2010).
22 Supplementary Information, Boucheron et al., Bilic, I. et al. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol 7, (2006). 4. Naoe, Y. et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med 204, (2007). 5. Yang, Y., Ochando, J.C., Bromberg, J.S. & Ding, Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood 110, (2007). 6. Sakaguchi, S. et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol 11, (2010). 7. Qui, H.Z. et al. CD134 plus CD137 dual costimulation induces Eomesodermin in CD4 T cells to program cytotoxic Th1 differentiation. J Immunol 187, (2011). 8. Cruz-Guilloty, F. et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 206, (2009). 9. Djuretic, I.M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol 8, (2007). 10. Sato, T. et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22, (2005). 11. Hassan, H. et al. Cd8 enhancer E8I and Runx factors regulate CD8alpha expression in activated CD8+ T cells. Proc Natl Acad Sci U S A 108, (2011).
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Nature Immunology: doi: /ni Supplementary Figure 1. Foxm1 loss leads to bone marrow hypocellularity.
 Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
High mobility group 1 HMG1
 Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
BD Cytometric Bead Array Flex Set System
 BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
FORMULAS FOR STATISTICS 1
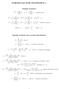 FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion
 Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks. Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks
 Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
[1] P Q. Fig. 3.1
![[1] P Q. Fig. 3.1 [1] P Q. Fig. 3.1](/thumbs/79/80362156.jpg) 1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
519.22(07.07) 78 : ( ) /.. ; c (07.07) , , 2008
 .. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
.. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo
 Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Aquinas College. Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET
 Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Figure 3 Three observations (Vp, Vs and density isosurfaces) intersecting in the PLF space. Solutions exist at the two indicated points.
 φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
J. Dairy Sci. 93: doi: /jds American Dairy Science Association, 2010.
 Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 651 APPENDIX B. BIBLIOGRAPHY 677 APPENDIX C. ANSWERS TO SELECTED EXERCISES 679
 APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Study on Re-adhesion control by monitoring excessive angular momentum in electric railway traction
 () () Study on e-adhesion control by monitoring excessive angular momentum in electric railway traction Takafumi Hara, Student Member, Takafumi Koseki, Member, Yutaka Tsukinokizawa, Non-member Abstract
() () Study on e-adhesion control by monitoring excessive angular momentum in electric railway traction Takafumi Hara, Student Member, Takafumi Koseki, Member, Yutaka Tsukinokizawa, Non-member Abstract
Identification of Fish Species using DNA Method
 5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
* ** *** *** Jun S HIMADA*, Kyoko O HSUMI**, Kazuhiko O HBA*** and Atsushi M ARUYAMA***
 J. Jpn. Soc. Soil Phys. No. +*2, p. +3,2,**2 * ** *** *** Influence Area of Stem Flow on a Soil of Deciduous Forest Floor by Electric Resistivity Survey and the Evaluation of Groundwater Recharge through
J. Jpn. Soc. Soil Phys. No. +*2, p. +3,2,**2 * ** *** *** Influence Area of Stem Flow on a Soil of Deciduous Forest Floor by Electric Resistivity Survey and the Evaluation of Groundwater Recharge through
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
,,, (, 100875) 1989 12 25 1990 2 23, - 2-4 ;,,, ; -
 25 3 2003 5 RESOURCES SCIENCE Vol. 25 No. 3 May 2003 ( 100875) : 500L - 2-4 - 6-8 - 10 114h - 120h 6h 1989 12 25 1990 2 23-2 - 4 : ; ; - 4 1186cm d - 1 10cm 514d ; : 714 13 317 714 119 317 : ; ; ; :P731
25 3 2003 5 RESOURCES SCIENCE Vol. 25 No. 3 May 2003 ( 100875) : 500L - 2-4 - 6-8 - 10 114h - 120h 6h 1989 12 25 1990 2 23-2 - 4 : ; ; - 4 1186cm d - 1 10cm 514d ; : 714 13 317 714 119 317 : ; ; ; :P731
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,
![[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13, [11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,](/thumbs/89/98694254.jpg) in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
(1) Describe the process by which mercury atoms become excited in a fluorescent tube (3)
 Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Buried Markov Model Pairwise
 Buried Markov Model 1 2 2 HMM Buried Markov Model J. Bilmes Buried Markov Model Pairwise 0.6 0.6 1.3 Structuring Model for Speech Recognition using Buried Markov Model Takayuki Yamamoto, 1 Tetsuya Takiguchi
Buried Markov Model 1 2 2 HMM Buried Markov Model J. Bilmes Buried Markov Model Pairwise 0.6 0.6 1.3 Structuring Model for Speech Recognition using Buried Markov Model Takayuki Yamamoto, 1 Tetsuya Takiguchi
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
ITU-R BT ITU-R BT ( ) ITU-T J.61 (
 ITU-R BT.439- ITU-R BT.439- (26-2). ( ( ( ITU-T J.6 ( ITU-T J.6 ( ( 2 2 2 3 ITU-R BT.439-2 4 3 4 K : 5. ITU-R BT.24 :. ITU-T J.6. : T u ( ) () (S + L = M) :A :B :C : D :E :F :G :H :J :K :L :M :S :Tsy :Tlb
ITU-R BT.439- ITU-R BT.439- (26-2). ( ( ( ITU-T J.6 ( ITU-T J.6 ( ( 2 2 2 3 ITU-R BT.439-2 4 3 4 K : 5. ITU-R BT.24 :. ITU-T J.6. : T u ( ) () (S + L = M) :A :B :C : D :E :F :G :H :J :K :L :M :S :Tsy :Tlb
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
Comparison of Evapotranspiration between Indigenous Vegetation and Invading Vegetation in a Bog
 J. Jpn. Soc. Soil Phys. No. +*-, p.-3.1,**0 ** * *** Comparison of Evapotranspiration between Indigenous Vegetation and Invading Vegetation in a Bog Toshiki FUJIMOTO*, Ippei IIYAMA*, Mai SAKAI*, Osamu
J. Jpn. Soc. Soil Phys. No. +*-, p.-3.1,**0 ** * *** Comparison of Evapotranspiration between Indigenous Vegetation and Invading Vegetation in a Bog Toshiki FUJIMOTO*, Ippei IIYAMA*, Mai SAKAI*, Osamu
Supplemental file 3. All 306 mapped IDs collected by IPA program. Supplemental file 6. The functions and main focused genes in each network.
 LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
UMI Number: All rights reserved
 UMI Number: 3408360 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send
UMI Number: 3408360 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplementary Information Titles
 Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά
 ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ ΝΟΣΗΛΕΥΤΙΚΗ 2008, 47(3):367 373 H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά Ανθή Χρυσοστόμου Νοσηλεύτρια ΠΕ, ΜSc, Kέντρο Ελέγχου και Πρόληψης
ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ ΝΟΣΗΛΕΥΤΙΚΗ 2008, 47(3):367 373 H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά Ανθή Χρυσοστόμου Νοσηλεύτρια ΠΕ, ΜSc, Kέντρο Ελέγχου και Πρόληψης
Supporting information. Influence of Aerosol Acidity on the Chemical Composition of Secondary Organic Aerosol from β caryophyllene
 Supporting information Influence of Aerosol Acidity on the Chemical Composition of Secondary Organic Aerosol from β caryophyllene M. N. Chan 1, J. D. Surratt 2,*, A. W. H. Chan 2,**, K. Schilling 2, J.
Supporting information Influence of Aerosol Acidity on the Chemical Composition of Secondary Organic Aerosol from β caryophyllene M. N. Chan 1, J. D. Surratt 2,*, A. W. H. Chan 2,**, K. Schilling 2, J.
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
A Bonus-Malus System as a Markov Set-Chain. Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics
 A Bonus-Malus System as a Markov Set-Chain Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics Contents 1. Markov set-chain 2. Model of bonus-malus system 3. Example 4. Conclusions
A Bonus-Malus System as a Markov Set-Chain Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics Contents 1. Markov set-chain 2. Model of bonus-malus system 3. Example 4. Conclusions
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov.
 A. ΈΛΕΓΧΟΣ ΚΑΝΟΝΙΚΟΤΗΤΑΣ A 1. Έλεγχος κανονικότητας Kolmogorov-Smirnov. Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov. Μηδενική υπόθεση:
A. ΈΛΕΓΧΟΣ ΚΑΝΟΝΙΚΟΤΗΤΑΣ A 1. Έλεγχος κανονικότητας Kolmogorov-Smirnov. Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov. Μηδενική υπόθεση:
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
Table of Contents 1 Supplementary Data MCD
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2017 Supporting Information for Magnetic circular dichroism and density functional theory
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2017 Supporting Information for Magnetic circular dichroism and density functional theory
EE512: Error Control Coding
 EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
Electronic Supplementary Information
 Electronic Supplementary Information NbCl 3 -catalyzed [2+2+2] intermolecular cycloaddition of alkynes and alkenes to 1,3-cyclohexadiene derivatives Yasushi Obora,* Keisuke Takeshita and Yasutaka Ishii*
Electronic Supplementary Information NbCl 3 -catalyzed [2+2+2] intermolecular cycloaddition of alkynes and alkenes to 1,3-cyclohexadiene derivatives Yasushi Obora,* Keisuke Takeshita and Yasutaka Ishii*
a 2,5 b 2,5 upplemental Figure 1 IL4 (4h) in Ja18-/- mice IFN-γ (16h) in Ja18-/- mice 1,5 1,5 ng/ml ng/ml 0,5 0,5 4ClPh PyrC 4ClPh PyrC
 a 2,5 IL4 (4h) in Ja18-/- mice b 2,5 IFN-γ (16h) in Ja18-/- mice 2 2 ng/ml 1,5 ng/ml 1,5 1 1,5,5 4ClPh NC PyrC 4ClPh NC PyrC upplemental Figure 1 a b c PyrC-α-GalCer NU-α-GalCer α-galcer CD4 (PE) MFI CD4
a 2,5 IL4 (4h) in Ja18-/- mice b 2,5 IFN-γ (16h) in Ja18-/- mice 2 2 ng/ml 1,5 ng/ml 1,5 1 1,5,5 4ClPh NC PyrC 4ClPh NC PyrC upplemental Figure 1 a b c PyrC-α-GalCer NU-α-GalCer α-galcer CD4 (PE) MFI CD4
DuPont Suva. DuPont. Thermodynamic Properties of. Refrigerant (R-410A) Technical Information. refrigerants T-410A ENG
 Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Electronic Supplementary Information
 Electronic Supplementary Information The preferred all-gauche conformations in 3-fluoro-1,2-propanediol Laize A. F. Andrade, a Josué M. Silla, a Claudimar J. Duarte, b Roberto Rittner, b Matheus P. Freitas*,a
Electronic Supplementary Information The preferred all-gauche conformations in 3-fluoro-1,2-propanediol Laize A. F. Andrade, a Josué M. Silla, a Claudimar J. Duarte, b Roberto Rittner, b Matheus P. Freitas*,a
Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil
 J. Jpn. Soc. Soil Phys. No. +*0, p.- +*,**1 Eh * ** Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil Daisuke MURAKAMI* and Tatsuaki KASUBUCHI** * The United Graduate
J. Jpn. Soc. Soil Phys. No. +*0, p.- +*,**1 Eh * ** Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil Daisuke MURAKAMI* and Tatsuaki KASUBUCHI** * The United Graduate
SUPPLEMENTARY INFORMATION
 Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Inverse trigonometric functions & General Solution of Trigonometric Equations. ------------------ ----------------------------- -----------------
 Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Inverse trigonometric functions & General Solution of Trigonometric Equations. 1. Sin ( ) = a) b) c) d) Ans b. Solution : Method 1. Ans a: 17 > 1 a) is rejected. w.k.t Sin ( sin ) = d is rejected. If sin
Supporting Information
 Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp.
 SUPPLEMENTARY MATERIAL New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp. Mohamed-Elamir F. Hegazy a,*, Amira M. Gamal-Eldeen b, Tarik A. Mohamed a, Montaser A. Alhammady c, Abuzeid A.
SUPPLEMENTARY MATERIAL New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp. Mohamed-Elamir F. Hegazy a,*, Amira M. Gamal-Eldeen b, Tarik A. Mohamed a, Montaser A. Alhammady c, Abuzeid A.
c Key words: cultivation of blood, two-sets blood culture, detection rate of germ Vol. 18 No
 2008 245 2 1) 1) 2) 3) 4) 1) 1) 1) 1) 1), 2) 1) 2) 3) / 4) 20 3 24 20 8 18 2001 2 2 2004 2 59.0 2002 1 2004 12 3 2 22.1 1 14.0 (CNS), Bacillus c 2 p 0.01 2 1 31.3 41.9 21.4 1 2 80 CNS 2 1 74.3 2 Key words:
2008 245 2 1) 1) 2) 3) 4) 1) 1) 1) 1) 1), 2) 1) 2) 3) / 4) 20 3 24 20 8 18 2001 2 2 2004 2 59.0 2002 1 2004 12 3 2 22.1 1 14.0 (CNS), Bacillus c 2 p 0.01 2 1 31.3 41.9 21.4 1 2 80 CNS 2 1 74.3 2 Key words:
DuPont Suva 95 Refrigerant
 Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
An experimental and theoretical study of the gas phase kinetics of atomic chlorine reactions with CH 3 NH 2, (CH 3 ) 2 NH, and (CH 3 ) 3 N
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 An experimental and theoretical study of the gas phase kinetics of atomic chlorine
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 An experimental and theoretical study of the gas phase kinetics of atomic chlorine
Daewoo Technopark A-403, Dodang-dong, Wonmi-gu, Bucheon-city, Gyeonggido, Korea LM-80 Test Report
 LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
CorV CVAC. CorV TU317. 1
 30 8 JOURNAL OF VIBRATION AND SHOCK Vol. 30 No. 8 2011 1 2 1 2 2 1. 100044 2. 361005 TU317. 1 A Structural damage detection method based on correlation function analysis of vibration measurement data LEI
30 8 JOURNAL OF VIBRATION AND SHOCK Vol. 30 No. 8 2011 1 2 1 2 2 1. 100044 2. 361005 TU317. 1 A Structural damage detection method based on correlation function analysis of vibration measurement data LEI
«Συντήρηση αχλαδιών σε νερό. υπό την παρουσία σπόρων σιναπιού (Sinapis arvensis).»
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
Supplement of Using empirical orthogonal functions derived from remote-sensing reflectance for the prediction of phytoplankton pigment concentrations
 Supplement of Ocean Sci., 11, 139 158, 2015 http://www.ocean-sci.net/11/139/2015/ doi:10.5194/os-11-139-2015-supplement Author(s) 2015. CC Attribution 3.0 License. Supplement of Using empirical orthogonal
Supplement of Ocean Sci., 11, 139 158, 2015 http://www.ocean-sci.net/11/139/2015/ doi:10.5194/os-11-139-2015-supplement Author(s) 2015. CC Attribution 3.0 License. Supplement of Using empirical orthogonal
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS
 CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
HIV HIV HIV HIV AIDS 3 :.1 /-,**1 +332
 ,**1 The Japanese Society for AIDS Research The Journal of AIDS Research +,, +,, +,, + -. / 0 1 +, -. / 0 1 : :,**- +,**. 1..+ - : +** 22 HIV AIDS HIV HIV AIDS : HIV AIDS HIV :HIV AIDS 3 :.1 /-,**1 HIV
,**1 The Japanese Society for AIDS Research The Journal of AIDS Research +,, +,, +,, + -. / 0 1 +, -. / 0 1 : :,**- +,**. 1..+ - : +** 22 HIV AIDS HIV HIV AIDS : HIV AIDS HIV :HIV AIDS 3 :.1 /-,**1 HIV
Mouse Gene 2.0 ST Array Shn3. Runx2 Shn3. Runx2 Shn3 Runx2. adult. (Schnurri, Shn)3/Hivep3. Runx2 Shn/Hivep. ST2 BMP-2 Shn3
 adult (Schnurri, Shn)3/Hivep3 Runx2 Shn/Hivep Shn3 Shn3 Shn3 Shn3 Shn1 Shn2 Shn3 gain-of-function loss-of-function Shn3 in vivo mimic MC3T3-E1 ST-2 ATDC5 C3H10T1/2 ALP RT-PCR alcian blue RT-PCR Affymetrix
adult (Schnurri, Shn)3/Hivep3 Runx2 Shn/Hivep Shn3 Shn3 Shn3 Shn3 Shn1 Shn2 Shn3 gain-of-function loss-of-function Shn3 in vivo mimic MC3T3-E1 ST-2 ATDC5 C3H10T1/2 ALP RT-PCR alcian blue RT-PCR Affymetrix
The toxicity of three chitin synthesis inhibitors to Calliptamus italicus Othoptera Acridoidea
 2011 48 4 909 914 * 1 2** 2 2 2 2 2*** 1. 110161 2. 100081 026000 3 Calliptamus italicus L. 3 LC 50 LC 90 1. 34 14. 17 mg / L LC 50 LC 90 2. 09 45. 22 mg / L 50 mg / L 14 d 87% 100% 50 mg / L 50% The toxicity
2011 48 4 909 914 * 1 2** 2 2 2 2 2*** 1. 110161 2. 100081 026000 3 Calliptamus italicus L. 3 LC 50 LC 90 1. 34 14. 17 mg / L LC 50 LC 90 2. 09 45. 22 mg / L 50 mg / L 14 d 87% 100% 50 mg / L 50% The toxicity
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Si + Al Mg Fe + Mn +Ni Ca rim Ca p.f.u
 .6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
.6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
