Supplementary Appendix; Figures S1-S16.
|
|
|
- Οὐρβανός Βαρουξής
- 6 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and polarised under Th1,, Th9, Th17 and itreg conditions in-vitro. Either all cells (Bulk) or FACS-purified Ifnγ yfp+, Il4 gfp+, Il9 Cre R26 eyfp+ Il17a Cre R26 efp635 reporter positive cells were isolated, RNA extracted and used for mirna and mrna transcriptional analysis. The mean of 3-5 biological replicates per T cell subset are represented in each heat map. a, Hallmark Th and Treg genes are shown. b, Venn diagrams (top) depicting numbers of common and exclusive mrna and mirnas expressed in bulk and purified cells (2-fold change). Bottom: Heat maps showing expression of mrna and mirnas in cells, relative to un-polarised cells. Supplementary Appendix; Figure S2. FACS-Purified reporter + T cells reveal significant IL-4-signaling resolution. CD4 T cells were purified and polarised under Th1,, Th9, Th17 and itreg conditions in vitro, using cytokine reporter cells. Either all cells (Bulk) or FACS-purified Ifnγ yfp+, Il4 gfp+, Il9 Cre R26 eyfp+ Il17a Cre R26 efp635 reporter positive cells were isolated (a). Using Ingenuity pathways analysis, the IL-4 signaling pathway was overlaid with the gene expression profile of bulk, or cytokinereporter purified cells (b, c). Supplementary Appendix; Figure S3. Naïve CD4 T cells were polarised in vitro, as described in methods, with mrna and mirna transcriptional profiles used for comparative analysis. a, Comparative analysis showing common and unique mrna (left) and mirna (right) transcripts in each T cell subset (>5 fold-change). Unique transcripts in cells highlighted in red square. b, Selection of -enriched mrnas (left) and mirnas (right). Supplementary Appendix; Figure S4. Fold change filters identify more abundant, and putatively more specific in vitro transcripts. Comparative analysis showing common and unique mrna (top) and mirna (bottom) transcripts in each in vitro generated T cell subset, as in Figure 1, (>2 fold-change, top two 1
2 tables, >10 fold-change, lower two tables). Unique transcripts in cells in red highlighted square. Supplementary Appendix; Figure S5. General down-regulation of mirnas and mirna processing pathways in cytokine + or TF + cells. Fold change of all mirnas, relative to naïve T cells, identified in in vitro and ex vivo cells, showing the mean (average) and frequency of up-regulated and down-regulated mirnas (a). Expression mirna biogenesis, processing and functional partners in all T cell samples in vitro and ex vivo (b). Supplementary Appendix; Figure S6. Fold change filters identify more abundant, and putatively more specific ex vivo transcripts. Comparative analysis showing common and unique mrna (top) and mirna (bottom) transcripts in each ex vivo isolated T cell subset, as in Figure 2, (>2 fold-change, top two tables, >10 fold-change, lower two tables). Unique transcripts in cells in red highlighted square. Supplementary Appendix; Figure S7. Transcriptional analysis of FACS-purified reporter + T cells reveals diversity between populations. From 4-way Venn diagrams showing comparative analysis of common and unique mrna (a) and mirna transcripts (b) between subsets (as depicted in Figure 1i). In a and b, left, total number of unique (only expressed in one sample), commonly expressed in two samples (pair, middle) and transcripts differentially expressed in 3 or more samples (cluster, right). Supplementary Appendix; Figure S8. Increasing fold change filters identify more abundant, and putatively more specific Heligmosomoides polygyrus 1 o - elicited transcripts. Comparative analysis showing common and unique mrna (top) and mirna (bottom) transcripts in each ex vivo isolated T cell subset, as in Figure 2, (>2 fold-change, upper left tables; >5 fold-change, lower left tables >10 fold-change, upper right two tables). Unique transcripts in cells in red highlighted square. Supplementary Appendix; Figure S9. Increasing fold change filters identify more abundant, and putatively more specific Heligmosomoides polygyrus 2 o - elicited transcripts. Comparative analysis showing common and unique mrna 2
3 (top) and mirna (bottom) transcripts in each ex vivo isolated T cell subset, as in Figure 2, (>2 fold-change, upper left tables; >5 fold-change, lower left tables >10 fold-change, upper right two tables). Unique transcripts in cells in red highlighted square. Supplementary Appendix; Figure S10. Four way comparative analyses identifies 82 common mrna and 14 common mirna transcripts in ex vivo and in vitro-derived cells. Using a 4-way comparative analysis on gene expression profiles from samples, 82 commonly expressed mrna transcripts (a) and 14 commonly expressed mirnas (b) were identified. Supplementary Appendix; Figure S11. Expression pairing using -enriched mrna and mirna transcripts identifies distinct candidate mirnas from in vitro and ex vivo cells. a-d, Using -enriched gene lists, as described in figure 2, expression-pairing analysis (i.e. elevated mirnas with predicted mrna targets that are significantly down regulated, and vice versa) shown at various fold change filters for in vitro derived (a), HDM-induced (b), H.p.1 o (c) and H.p. 2 o (d) cells. Bar charts on right showing the frequency of mrna targets identified for each mirna. e, Total number of mrnas targeted and inversely expressed in each sample by each candidate mirna. f, Expression of candidate mirnas in each sample, relative to naïve T cells. Supplementary Appendix; Figure S12. Dynamic expression of candidate mirnas and their mrna targets in vitro and ex vivo. Naïve T cells were cultured under -polarising conditions. At indicated days post-polarization, IL4 gfp+ cells were FACS purified for RNA analysis. a, Expression of candidate mirnas (top and middle, highlighting day 7 expression levels) and -associated genes (bottom), expressed as relative fold change to RNU6b or HPRT, as indicated. b, Expression of selected mrna targets of each candidate mirna at indicated times post polarisation. c, Expression of mir-155, mir-146a and their target genes in cells isolated from the airspaces (bronchoalveolar lavage fluid (BAL)) or local thoracic lymph nodes (tln) of HDM-challenged mice. Data expressed relative to RNU6b or HPRT. 3
4 Supplementary Appendix; Figure S13. Four way comparative analyses of mrna targets of selected mirna candidates, across all 4 samples. Left, We used a 4-way analysis, depicted as a 4-way Venn diagram, to identify mirnas, which target the same mrna in each subset (a-e). For example, mir- 146a targeted 44 mrnas in HDM cells, but none of these mrnas were predicted targets of mir-146a in other samples. In contrast mir-146a was predicted to target, and had inverse expression, with the same 14 mrnas in HDM cells and both H. polygyrus-elicited cells. Right, Selected mrna targets and expression in each subset, relative to naïve T cells. Fold change is relative to naïve CD4 + T cells. Supplementary Appendix; Figure S14. Baseline Th and Treg cell responses in chimera systems. a, Schematic representation of the generation of mir-146a mixed T cell bone marrow chimeric mice. b. Frequency of CD4 + CD25 + Foxp3 + cells in the spleen of naïve chimeric mice after 6-8 weeks of re-constitution. c. Frequency of Th1 (IFNγ + ), (IL-5 + or IL-13 + ) and Th17 (IL-17 + ) cells in the spleen of naïve chimeric mice after 6-8 weeks of re-constitution. d. Schematic representation of the generation of mir-155 mixed T cell bone marrow chimeric mice. e. Frequency of CD4 + CD25 + Foxp3 + cells in the spleen of naïve chimeric mice after 6-8 weeks of re-constitution. f. Frequency of Th1 (IFNγ + ), (IL-5 + or IL-13 + ) and Th17 (IL-17 + ) cells in the spleen of naïve chimeric mice after 6-8 weeks of re-constitution. Supplementary Appendix; Figure S15. mir-155 is not required for anti-malarial Th1 response. WT and mir-155 / mice were infected (i.p.) with 10 5 P. chabaudi infected RBCs. a, Total splenocytes were counted at day 8 following infection. b, Whole blood was recovered with hematology assessed using a vetscan system. c, Splenic Th1 cells were assessed following PMA and ionomycin stimulation and intracellular cytokine staining. d, The percentage of infected red blood cells were enumerated on fresh blood smears. Supplementary Appendix; Figure S16. S1pr1 sirna. a, Naïve WT T cells were treated with S1pr1 sirna (nm) or with scrambled control sirna. Cells were incubated for 24 hours before S1pr1 was determined by qrt-pcr. 4
5 Supplementary Appendix; Figure S1. Fold change (relative to naive CD4 + ) a Fold change (relative to naive CD4 + ) Fold change (relative to naive CD4 + ) Fold change (relative to naive CD4 + ) Fold change (relative to naive CD4 + ) IL-10 GZMB GZMA IL IL-2 IL-21 IL-2 IL-21 IFNg IL-2 SOCS2 CD25 Th1 IL-10 IL-21 GZMB CD25 TNF IL-4 LIF IL-24 IL-2 Th17 Tbx21 CCR1 IL-17A CyP1A1 BHLHE40 IL-22 CYP1B1 IL-1R2 Foxp3 CTLA4 RRM2 NRP1 (Neuropilin) Th9 IL-10 IL-4 IL-17A CD25 IL-9 IL-24 BHLHE40 itreg b AREG SOCS2 E4BP4 RORa RORc CD44 CXCR6 CD69 IL-12Rb2 FOSB CD25 E4BP4 IRF4 AREG GATA3 IRF4 E4BP4 GATA3 RORa IKZF2 (Helios) TNFRSF4 (OX40) IL-2Rb CD38 ITGAE (CD103) IKZF4 (EOS) Bulk Purified Bulk CD4 + : mrna (2- fold) Purified Bulk polarised 3430 Naïve (CD4 + CD44 CD25 CD62L hi ) Purified polarised (CD4 + IL- 4 gfp+ ) Bulk CD4 + 8 Bulk : mirna (2- fold) polarised Naïve (CD4 + CD44 CD25 CD62L hi ) Purified Purified (CD4 + IL- 4 gfp+ )
6 Supplementary Appendix; Figure S2 a. c. Th1 Th9 Th17 itreg Ifnγ yfp Il4 gfp Il9 Cre R26 eyfp Il17 Cre R26 efp635 Foxp3 rfp Symbol IL-4 signaling pathway (Bulk) (Purified) Fold change Type(s) Entrez ID IL cytokine IRF transcription regulator SOCS other JAK kinase JAK kinase IL4R transmembrane receptor NFATC transcription regulator NFAT transcription regulator INPPL phosphatase RRAS enzyme AKT kinase NRAS enzyme NFATC transcription regulator RRAS enzyme KRAS enzyme FCER transmembrane receptor NR3C ligand-dependent nuclear receptor ATM kinase PIK3CD kinase PIK3R kinase PTPN phosphatase SOS other AKT kinase RPS6KB kinase PIK3R kinase b. (Bulk) (CD4 + IL- 4gfp + Purified)
7 Supplementary Appendix; Figure S3 In- vitro purified T cell subsets a mrna (5- fold) Th1 Th9 Th17 Treg mirna (5- fold) Th1 Th9 Th17 Treg b RNPC3 BTLA CREBZF CCR7 DICER1 RPRD1A Rassf1 CLCN3 LY96 TMEM45A CD99 CCRL2 JUNB IL4R PPARG JAK3 NFKBIZ CCR4 TGIF2 TM6SF1 JUN BATF TNFSF13B IL10RA SOCS1 JDP2 CCR2 KLF4 FOSB CCR1 Th Th Th Treg mrna enriched genes Fold change Th Th Th Treg mmu-mir-146a mmu-mir-200c mmu-mir-1839 mmu-mir-28 mmu-mir-15a mmu-mir-339 mmu-mir-324 mmu-mir-93* mmu-mir-345 mmu-mir-17* mmu-mir-330* mmu-mir-674 mmu-mir-1894 mmu-mir-468 mmu-mir-465c-2 mirna enriched mirna mmu-mir-2137 mmu-mir-3072* mmu-mir-667 mmu-mir-574 mmu-mir-682 mmu-mir-574* mmu-mir-2861 mmu-mir-466i mmu-mir-2182 mmu-mir-6690 mmu-mir-2137 mmu-mir-714 mmu-mir-760 mmu-mir-1224 mmu-mir-3470b mmu-mir-1892 mmu-mir-92a* mmu-mir-705 mmu-mir-1982* mmu-mir-16* mmu-mir-762 mmu-mir-1893 mmu-mir Fold change
8 Supplementary Appendix; Figure S4. In- vitro purified T cell subsets 2- fold change mrna (2- fold) Th1 Th9 Th17 Treg mrna Th Th Th itreg mirna (2- fold) Th1 Th9 Th17 Treg Th mirna Th Th itreg mrna (10- fold) 10- fold change Th1 Th9 Th17 Treg mrna Th Th Th itreg mirna (10- fold) Th1 Th9 Th17 Treg Th mirna Th Th itreg
9 Eif2c3 (Ago3) Eif2c2 (Ago2) Eif2c1 (Ago1) Supplementary Appendix; Figure S5 a In-vitro Th1 ( all mirnas) 1000 Ex-vivo Th1 ( all mirnas) Fold change (relative to naive CD4 + ) 10 1 Frequency = 46 Average Fold change (relative to naive CD4 + ) 10 1 Frequency = 70 Average 0.69 b 0.10 Frequency = Frequency = 75 Fold change (relative to naive CD4 + ) Fold change (relative to naive CD4 + ) 0.01 mirnas In-vitro ( all mirnas) mirnas In-vitro Th17 ( all mirnas) Frequency = 80 Average 0.33 Frequency = 86 Frequency = 56 Average Fold change (relative to naive CD4 + ) Fold change (relative to naive CD4 + ) 0.01 mirnas Ex-vivo (HDM) ( all mirnas) mirnas Average 0.76 Ex-vivo Th17 (EAE) (all mirnas) Frequency = 96 Frequency = 79 Frequency = 90 Average Fold change (relative to naive CD4 + ) Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 Th9 Th17 itreg Th1 Th9 Th17 itreg Th1 Th9 Th17 itreg Fold change (relative to naive CD4 + ) Dgcr8 Dicer Tarbp Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 Th9 Th17 itreg Th1 Th9 Th17 itreg mirnas Frequency = mirnas Frequency = 87 Eif2c4 (Ago4) Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 Th9 Th17 itreg In-vitro itreg (All mirnas) 1000 Ex-vivo ntreg (All mirnas) Th1 (P.c. (HDM) (H.p.1 o ) (H.p.2 o ) Th17 (EAE) ntreg Th1 Th9 Th17 itreg Fold change (relative to naive CD4 + ) 10 1 Frequency = 36 Average 0.56 Fold change (relative to naive CD4 + ) 1 Frequency = 102 Average Frequency = 56 Frequency = mirnas 0.01 mirnas
10 Supplementary Appendix; Figure S6 Ex- vivo isolated T cell subsets mrna mrna (2- fold) Th1 P.C HDM Th17 E.A.E. Th1 P.C 2- fold change HDM Th17 E.A.E. ntreg ntreg mirna mirna (2- fold) Th1 P.C HDM Th17 E.A.E. Th1 P.C HDM Th17 E.A.E. ntreg ntreg fold change mrna mrna (10- fold) Th1 P.C HDM Th17 E.A.E. Th1 P.C HDM Th17 E.A.E. ntreg ntreg mirna (10- fold) Th1 P.C HDM Th17 E.A.E. ntreg mirna Th1 P.C HDM Th17 E.A.E ntreg
11 Supplementary Appendix; Figure S7 Ex- vivo isolated subsets a No. of transcripts mrna unique pair cluster (3+) a b c d e f g h i j k l m n o HDM Hp 1o Hp 2o In- vitro b No. of transcripts unique pair cluster (3+) mirna a b c d e f g h i j k l m n o HDM Hp 1o Hp 2o In- vitro
12 Supplementary Appendix; Figure S8 Hp 1 o (- enriched mrna and mirna) 2- fold change 10- fold change mrna (2- fold) Th1 P.C Hp 1 o Th17 E.A.E. ntreg mrna (10- fold) Th1 P.C Hp 1 o Th17 E.A.E. ntreg Th1 P.C Th1 P.C mrna Hp 1 o Th17 E.A.E mrna Hp 1 o Th17 E.A.E ntreg ntreg mirna (2- fold) Th1 P.C Hp 1 o Th17 E.A.E. ntreg mirna (10- fold) Th1 P.C Hp 1 o Th17 E.A.E. ntreg mirna Th1 P.C Hp 1 o Th17 E.A.E mirna Th1 P.C Hp 1 o Th17 E.A.E ntreg ntreg fold change mrna (5- fold) Th1 P.C Hp 1 o Th17 E.A.E. ntreg mrna Th1 P.C Hp 1 o Th17 E.A.E ntreg mirna (5- fold) Th1 P.C Hp 1 o Th17 E.A.E. ntreg mirna Th1 P.C Hp 1 o Th17 E.A.E ntreg
13 Supplementary Appendix; Figure S9 Hp 2 o (- enriched mrna and mirna) 2- fold change 10- fold change mrna (2- fold) Th1 P.C Hp 2 o Th17 E.A.E. ntreg mrna (2- fold) Th1 P.C Hp 2 o Th17 E.A.E. ntreg Th1 P.C Th1 P.C mrna Hp 2 o Th17 E.A.E mrna Hp 2 o Th17 E.A.E ntreg ntreg mirna (2- fold) Th1 P.C Hp 2 o Th17 E.A.E. ntreg mirna (2- fold) Th1 P.C Hp 2 o Th17 E.A.E. ntreg mirna Th1 P.C Hp 2 o Th17 E.A.E mirna Th1 P.C Hp 2 o Th17 E.A.E ntreg ntreg fold change mrna (10- fold) Th1 P.C Hp 2 o Th17 E.A.E. ntreg mrna Th1 P.C Hp 2 o Th17 E.A.E ntreg mirna (10- fold) Th1 P.C Hp 2 o Th17 E.A.E. ntreg mirna Th1 P.C Hp 2 o Th17 E.A.E ntreg
14 Supplementary Appendix; Figure S10 a. mrna transcripts b. mirnas Atp1b1 A930005H10Rik Sox4 Cmah Ms4a4b Ovgp1 Ramp1 Nacc2 Pik3ip1 Dzip1 Vipr1 Adh1 Rapgef4 Mtss1 Xkrx Stk38 Pdk1 Angptl2 Cpd Cyfip1 Csda Eea1 Ptpn13 Plcb4 Cobll1 Slc39a4 Zdhhc2 Ahr Ttc39c B4galnt4 Fgl2 Rbpj Ptger2 Acot7 Pdcd1 Chst2 Il12rb1 Fam129a Chsy1 Cks2 Mdfic Il2rb Csf1 Ell2 Metrnl Ccr2 Lgals7 Hspa1b Tnfsf11 Cxcr6 Tbx21 Ccnb2 Maf Mki67 Stx11 Cish Tiam1 Casp4 Cass4 Cst7 Il1rl1 Pdcd1lg2 Rnf128 Gpm6b Ccl1 Penk Prelid2 Kcnk5 Ctla2a Prnp Rgs1 Socs2 Nfil3 Frmd4b Tnfsf14 Il2 Il1r2 Egln Fold chnage (relative toi naive CD4) HDM In-vitro Hp2o Hp1o mmu-mir-150-star mmu-mir-146a mmu-mir-200c mmu-mir-151-5p mmu-mir-378-star mmu-mir-151-3p mmu-mir-339-5p mmu-mir-874 mmu-mir-720 mmu-mir-30c-1-star mmu-mir-674 mmu-mir-1955 mmu-mir-182 mmu-mir Fold chnage (relative toi naive CD4) HDM In-vitro Hp2o Hp1o
15 Supplementary Appendix; Figure S11 a b c mirna&target&enrichment:&& In&vitro&5enriched&miRNAs&which&target&5enriched&genes&& mirna&& (fold&change&filter)& 25fold& 55fold& 105fold& mrna&& (fold&change&filter)& 25fold& 55fold& 105fold& mir%214%3p! mir515a& mir515a& Candidate&& mirnas&!!!!! mir%709! mir%28! mir520b& mir%760! mir5146a& mir%28! mir%762! mir5200c& mir5146a& mir%1224! mir%324! mir%185! mir%1893! mir%330*(3p)! mir5200c& mir%2861! mir%339! mir%221! mir%574! mir%324! mir%760*(3p)! mir%339! mir%1982*!(3p)! mir%1982*(3p)! mir%1839! mir%1839! mir%3941! mirna&target&enrichment:&& &(HDM)5enriched&miRNAs&which&target&&(HDM)5enriched&genes&& mirna&(fold&change&filter)& 25fold& 55fold& 105fold& mrna&(fold&change&filter)& 25fold& 55fold& 105fold& & mir57b& mir520a& mir522& Candidate&& mir51555hp& mir%18a! mirnas! mir%361! mir522&! mir%379! mir%494! mirna&target&enrichment:&& &(Hp&1 o )5enriched&miRNAs&which&target&&(Hp&1 o ))5enriched&genes&& mirna&& (fold&change&filter)& 25fold& 55fold& 105fold& mrna&& (fold&change&filter)& 25fold& 55fold& 105fold& Candidate&& mirnas& mir%140! None& None& mir%154! No. of mrna targets in enriched cells No. of mrna targets in enriched cells No. of mrna targets in enriched cells No. of mrna targets in enriched cells No. of mrna targets in enriched cells No. of mrna targets in enriched cells No. of mrna targets in enriched cells mir mir-7b mir-20a mir-22 mir-762 mir-2861 mir-709 mir-15a mir-20b mir-200c mir-146a mir-221 mir-185 mir-140 In vitro mir-155 mir-18a 2-fold (46 total) mir-760 mir-1893 mir fold mir-15a mir-200c mir-1982 mir-146a mir-324 mir-339 mir-1839 mir-3941 mir-574 mir-28 mir-760-3p mir-330-3p mir-379 mir fold mir-339 mir-28 mir-324 mir-1982* mir-1839 HDM-induced 2-fold 2-fold mir-154 mir fold mir fold Hp 1 o -induced d e mirna&target&enrichment:&& &(Hp&2 o )5enriched&miRNAs&which&target&&(Hp&2 o ))5enriched&genes&& mirna&& 25fold& 55fold& 105fold& (fold&change&filter)& HDM! mrna&& (fold&change&filter)& Candidate&& mirnas&! No.!of!differen,ally!! predicted!mrna!targets!!!! (Hp!1 o )!!! (Hp!2 o )! 25fold& 55fold& 105fold& None! MiR520b& None& InBvitro! mirb7b! 35# $# $# $# mirb15a!! 184# 186# 72# 206# mirb20a! 200## $# $# $# mirb20b! $# 98# 101# 215# mirb22! 60# $# $# 80# mirb140! $# 55# 7# 60# mirb146a! 44# 34# 18# 48# mirb154! $# 8# $# $# mirb155! 88# 77# 28# 119# mirb200c! 113# 77# 80# 151# f mir-146a ( Fold change relative to naive CD4 + ) in-vitro HDM Hp 1 o mir-200c (Fold change relative to naive CD4 + ) Hp 2 o in-vitro mir-15a (Fold change relative to naive CD4 + ) HDM Hp 1 o Hp 2 o No. of mrna targets in enriched cells in-vitro HDM Hp 2 o -induced mir-20b Hp 1 o mir-20b (Fold change relative to naive CD4 + ) Hp 2 o in-vitro HDM 5-fold mir-146a mir-15a mir-155 mir-200c mir-155 (Fold change relative to naive CD4 + ) Hp 1 o mir-20b 10 1 Hp 2 o in-vitro HDM Hp 1 o Hp 2 o
16 Supplementary Appendix; Figure S12 a Relative Fold change Days post polarisation Day 7 mir-146a mir-20b mir-15a mir-200c mir-155 mir-146a mir-15a mir-20b mir-155 mir-200c Stat6 Gata3 b Relative fold change 40 mir Sgk Il6r Rictor S1pr1 1.5 mir-15a Tia Cd2ap Foxo mir-146a Itch Stat Smad mir-20b Egr Hif1α Lif Rorα 1.5 mir-200c Abi Gfi Ell Ptpn13 mirna mirna targets Days post polarisation Il5 Il4 Il Tnfaip c 6 mir S1pr Rictor 100 Sgk3 100 Il6r days post polarisation Relative expression 4 2 relative expression relative expression relative expression 10 relative expression 10 0 BAL tln 0 BAL tln 1 BAL tln 1 BAL tln 1 BAL tln 0.15 mir-146a 100 Stat1 100 Itch 100 Smad4 Relative expression relative expression 10 relative expression 10 relative expression BAL tln 1 BAL tln 1 BAL tln 1 BAL tln
17 Supplementary Appendix; Figure S13 a in- vitro mir- 146a targets Hp 1 o HDM Hp 2 o Fold change (relative to naive CD4 + ) HDM Hp 1 o Hp 2 o In-vitro Itch Stat1 Smad4 b mir- 15a targets 0 Sgk3 Cd2ap Dennd1b -5 Foxo1 Nr2c2 c in- vitro Hp 1 o HDM Hp 2 o Fold change (relative to naive CD4 + ) HDM Hp 1 o Hp 2 o In-vitro Rictor Smad7 Tgfbr3 Tia1 Tsc1 mir- 155 targets in- vitro Hp 1 o HDM Hp 2 o Fold change (relative to naive CD4 + ) HDM Hp 1 o Hp 2 o In-vitro Sgk3 Il6R Rictor S1pr1 Tia1
18 Supplementary Appendix; Figure S13 40 Abi2 d 30 Ahnak Atxn1 Ell2 mir- 200c targets HDM Hp 2 o Fold change (relative to naive CD4 + ) Gfi1 Hipk2 Maf Ptpn in- vitro Hp 1 o 20 0 HDM Hp 1 o Hp 2 o In-vitro e mir- 20b targets Ahnak Egln3 Egr2 Fgl2 Hif1α in- vitro Hp 1 o Hp 2 o Fold change (relative to naive CD4 + ) Klf10 Lif Map3k8 Rnf128 (Grail) Rora Rps6ka5 Tnfaip3 Tnfsf11 (RankL) 0 HDM Hp 1 o Hp 2 o In-vitro
19 Supplementary Appendix; Figure S14. a 20% mir- 146a +/+ + 80% TCRα /! 20% mir- 146a / + 80% TCRα /! WT b Treg (CD4 + CD25 + Foxp + ) 25 c 25 Th1 (IFNγ + ) 25 (IL-5 + ) 25 (IL-13 + ) 25 Th17 (IL-17 + ) % of CD % of CD % of CD % of CD % of CD T (mir-146a +/+ ) T (mir-146a -/- ) 0 T (mir-146a +/+ ) T (mir-146a -/- ) 0 T (mir-146a +/+ ) T (mir-146a -/- ) 0 T (mir-146a +/+ ) T (mir-146a -/- ) 0 T (mir-146a +/+ ) T (mir-146a -/- ) d 20% mir /+ + 80% TCRα /! 20% mir- 155 / + 80% TCRα /! WT e Treg (CD4 + CD25 + Foxp + ) % of CD f % of CD Th1 (IFNγ + ) % of CD (IL-5 + ) % of CD (IL-13 + ) % of CD Th17 (IL-17 + ) 0 T (mir-155 +/+ ) T (mir-155 -/- ) 0 T (mir-155 +/+ ) T (mir-155 -/- ) 0 T (mir-155 +/+ ) T (mir-155 -/- ) 0 T (mir-155 +/+ ) T (mir-155 -/- ) 0 T (mir-155 +/+ ) T (mir-155 -/- )
20 Supplementary Appendix; Figure S15 a. b. Total Splenocytes RBC (Cells/L) n.s mir-155 +/+ mir-155 / 0 n.s. mir-155 +/+ mir-155 -/- Haemoglobin (g/dl) n.s. mir-155 +/+ mir-155 -/- Haematocrit (%) n.s. mir-155 +/+ mir-155 -/- Mean Corpuscular Volume (fl) n.s. mir-155 +/+ mir-155 -/- c. % CD44 + IFNγ IFNγ mir /+ mir- 155 / d. % infected RBC 0 mir-155 +/+ mir-155 / n.s. mir-155 +/+ mir-155 -/- CD44
21 Supplementary Appendix; Figure S16 a. S1pr1 (Reltaive expression) * Mock sirna S1pr1 sirna Scrambled
22 Supplementary Appendix; Tables 1-4. Supplementary Appendix; Tables 1. Gene expression data of bulk and purified cells. Bulk cells represent the whole well, while purified cells represent FACS purified IL4 gfp+ cells. Fold change relative to naïve (CD4 + CD44 CD62L hi CD25 cells). Data expressed as mean of 3 or 4 biological replicates. Supplementary Appendix; Tables 2. In-vitro -enriched genes using a >2fold cut off. Genes that were differentially regulated greater than or equal to 2 fold and were not differentially regulated in other T cell populations. Fold change relative to naïve (CD4 + CD44 CD62L hi CD25 cells). Data expressed as mean of 3 or 4 biological replicates. Supplementary Appendix; Tables 3. Ex-vivo -enriched mirnas using a 5-fold cut off. mirnas that were differentially regulated greater than or equal to 2, 5 or 10-fold and were not differentially regulated in other ex-vivo T cell populations. Fold change relative to naïve (CD4 + CD44 CD62L hi CD25 cells). Data expressed as mean of 3 or 4 biological replicates. Supplementary Appendix; Tables 3. Ex-vivo -enriched genes using a 5-fold cut off. Genes that were differentially regulated greater than or equal to 2, 5 or 10-fold and were not differentially regulated in other ex-vivo T cell populations. Fold change relative to naïve (CD4 + CD44 CD62L hi CD25 cells). Data expressed as mean of 3 or 4 biological replicates.
23 Symbol (Bulk) (purified) Fold change (relative to Naïve T cell) Entrez Gene ID for Mouse IL Gp49a/Lilrb GZMB GZMA IL IL4 (includes EG:16189) EGLN IL1R LIF IL ATF MT1E IL LGALS IER TNFSF FRMD4B CCR FOSB BHLHE EMP IL2RA TNFSF NFIL EG665536/Ifitm SOCS IRF RGS MT2A CCL Ctla2a MAP3K ECM1 (includes EG: ) AREG/AREGB PRNP PLSCR CTLA SERPINF CDKN1A HBEGF TNFRSF Trim GCH MAFF IFITM IL ARL5B FOSL DUSP
24 EGR SRXN KCNK NR4A CCR SEMA7A PRELID Gm PENK HK CCL S100A EFNA EMILIN ERRFI ANXA KLF TNFRSF GPM6B TNF RNF STC MCF2L J06Rik/LOC ERO1L Ctla2b ALDOC PDCD1LG ENTPD IL1RL ANXA RNF19B PHLDA UBE2C CST CASP SLAMF TIAM NUSAP RRM SLCO4A NDRG TRIB CISH AQP STX KCTD PLK RGS MKI ADAM C6orf ASNS
25 BIRC MGARP PLK MAF CCNB REL K04Rik SV2C EHD TBX GADD45B NFKBID SPRY KIAA NR4A SLAMF Crem CXCR SYTL Bnip CEP SDC GADD45G TNFSF SELENBP STC HSPA1A/HSPA1B LGALS7/LGALS7B DUSP CCNA CCR SERPINE ASPM SLA BUB METRNL KLF ELL LTB4R CSF IL2RB VLDLR TEX KRT MDFIC TUBE CKS PTPRK INHBA NR4A TOP2A PROS GPLD
26 EPAS F2R RILPL P4HA IGHG ACVR2A CDK PLXDC AI EBI CENPE PLA2G12A AURKB IL CENPF Ccnb1/Gm SERPINE SAMSN TGM CHSY FAM129A PLEK TPI PPP2R3A S100A CDCA CSRNP PBK TGIF CYSTM CST GFI CD40LG IL12RB EGR CTSE NKG MZB DUSP ICOS SPC CHST PDCD AKR1C C CDCA ADM IL18RAP ACSL IFITM HIPK SPAG ASB
27 JDP AK SLC41A CASP CDC EHD FOS ESM SKIL CCR FASLG F2RL GPRIN FFAR PIM SOCS HIF1A MMP ECT TMEM176A CXCR SAP TNFAIP IGHA C9orf PRC PLCD OIT SLC2A NEK PMEPA MST1R SFT2D NUPR ITGA GPR EGR ACOT GNA TK TTC39B MYO BCL2A PTGER PSRC RUNX ODC CDCA NCAPG GCM AGPAT PLP FAM132A
28 FABP ADAMTSL GABPB GLIPR DCUN1D AHNAK CAMK2N Pmaip RBPJ CD8A IL10RA HLA-A HSPA4L LGALS TACC CACNB mir CDK TUBB CSRP IGHM PRDM LAMA NFKBIE Serpina3g (includes others) MAPKAPK DIAPH RACGAP RAD54L BUB1B TNFSF13B ORC ATL HILPDA FAM107B FGL MYO1F TRAF B4GALNT SYCE MGLL TMEM176B PLAC FURIN BZRAP CXCR ZC3H12C MAP2K KIF20A TTC39C MYC TPX PYGL
29 CD AHR EVI2A ERMN UHRF MXD TG GLRX SPC BATF GOT CASP STARD JUN BASP OSBPL GBP CKS1B FADS MLLT EIF4EBP DUSP BRCA RAD C13orf ZC3H12A SEPT RNF JAK KIF3A CD MELK ZDHHC TRIP TYMS SLC39A ETV MIS18BP AHCYL ISG PIWIL HSD17B GNA CHST Gm TM6SF NEDD RNF19A MYADM GABARAPL PKIG PPP1R15A CENPK
30 TIPARP RNF CASP OBFC2A MARCKSL MAPRE DNAJB RPS6KA VEGFA STIL RAD51AP RSAD CKAP SLC4A BTG LMNA BCL2L SOAT SHROOM FIGNL NFIA ID KIF4A KNTC TMEM GZMK VIM HMMR RRAGD COBLL FES CD E2F GATA N4BP PLCB AURKA TNFSF TANC CLDND SLC39A LAG RLIM CCL ESPL FAM110A BCAT CLSPN ORAI CCDC CDC S100A HIP
31 HEMK KDM6B ENPP CD SESN PTPN CTTN TGIF EEA CFLAR CSDA KLRC ZNF ARHGAP C21orf TRPM SDF CCR PIM FOCAD GINS CDKN PIK3AP AUH TNFRSF PGAM CHAC HGF ITGB SH2D2A ANLN ATXN7L NFKBIZ SCPEP ICAM GPR NEDD CLCN CLIC NCAPH PPP3CC MLKL GNG DUSP MSC JAK TMEM PKM ATP2B S100A GLA IMPA ALDOA
32 CAPG Gapdh (includes others) ARRDC Med NAMPT CRK MYO1C PTPN ARIH FAM46A HIST1H2AB/HIST1H2AE L24Rik Cd24a ZFX PLK PPARG CITED CARHSP PC PAG CENPH CA PGK IL4R NEIL LPXN GPT MED OPTN NFKB SDF2L ENC RHOQ TSC22D IRF MAPK FOXM GPR DGAT CYFIP JUNB SPTY2D IFRD XBP USP LXN H1F NCOA DTL ST3GAL ING CGGBP CENPP
33 CCRL COQ10B ANXA PRDX EZH MED LMAN TMEM SLC2A EPS ABI GRN ALCAM ZAK CSRP ATG9B NET Cd99 (mouse) CPD MCM10 (includes EG:307126) TMEM45A MBNL AFF DAPP CCRN4L CD LAMC IRS CENPN NUCB LYST CKB SCD ZNF LARGE MMS22L FNIP PTGER DDX KIF CDH RND GOLPH DKKL BMPR1A NRN LAMP SGOL GEMIN MARCKS CCDC109B LY GPNMB
34 CD CDC6 (includes EG:23834) CHD ARHGAP ITK ANGPTL IL23A Ly6a (includes others) ARHGAP11A CREB3L GSTK RRM SIAH SMOX CYP51A OXSM FNBP HELLS LRP DAPK PRR SLC37A SEC61A RALGDS FBXO CDC25C SERINC CHD NCKAP PFKP WASL LITAF PPAP2C GALK CEBPB UNC13A TWF KIAA ACSBG ACPP CDCA GNAQ INSIG CTNNA PPP2CB FHL SLC39A CLCN ATP9A SMAD LIG AIM GLS
35 ANKRD Scd HLA-DMA DYRK FAIM STK BCL KIF2C APBB PTPRV TMEM EMR PLK ARF VMP ITPR CCDC TSPAN KPNA NCAPD TLCD RCAN NUCB ASF1B TSPAN XCL A630033H20Rik DUSP GKAP SYNE HSPA ATF DUSP DNAJB BCL2L BSG UBL POLA SETD C9orf NFKBIB LONRF CCL PAFAH1B CAMK2D WISP ME Tagap ACAT ENDOD SLC16A NFATC GM2A
36 CENPM RNF FUT DOCK ZFP36L PGM H2-Q NFKBIA Rrbp Nup HMGCR ST8SIA Tcrg-V TJP HMOX KLF SURF TP53INP SDR39U CORO2A ADAM CD INCENP TFRC Zfp COX7A WHSC AGPAT SMC HMGB ARID5B TMEM SKAP ETF RORA GMDS DCXR HCCS FEM1C STK32C CD SQLE EPCAM PTPN LMNB BCL SLC25A FDPS FBXO PFKL TNFRSF1B MEFV FEM1B
37 ZFP RELB PLOD AFG3L BBX BARD ZBP CTSF EVI RRAD CYB DST ARFGAP LCP ARF HMGN HHEX MYL DDC SNAP PBX CCDC PFN MAN1A ZNF ATXN DMWD C17orf DEPDC1B Timd SRGN CKAP TSPAN TLK ZCCHC ZWINT GFPT ARL PLAGL GPC HMGCS TOLLIP TEP TMEM MTMR TRAF PTPN CIT PDCD6IP RDH SORD HIST1H3A (includes others) SLC31A
38 BCL ID MMP MCM SLC25A CDKN2C STX ADAM MCM Gm9790/Higd1a RYK POU2AF KCNQ FAR J17Rik FNDC3B HRH ABCC IPMK MCL SEC24D GMFB ATP2B GNG MAFG CXADR PLEKHA UCP RAPH MAP2K CYSLTR IL1R GORASP PRKRA COL18A MID1IP PRR GCNT LAT AXL POLE GPI SNX ZNF SYNGR MTMR LGALSL SWAP ATF LGALS3BP MAP CSNK1D DCP2 (includes EG:167227)
39 SLC35B IKZF SLC3A C15orf PGLYRP HPSE GCAT TASP ABHD GSN UGT1A NUDT PIP5K1B APLP GOSR ITGAV GRHPR DERL CLPB GRINA LOC RNF GINS NOL ARL4A EIF1AY SLC7A VTI1A SUCO ERCC6L ARIH GBP2 (includes EG:14469) DNAJC MAP RAB1A CNOT HLA-DOA C11orf PURB P2RX HSPA ARHGAP NSMCE UBE2S GPR PILRB MBOAT IQGAP Cyp4f16/Gm NCOA DCTN CHCHD GMNN
40 NEK VPS54 (includes EG:245944) P4HA FAM162A Gm SSR ANG MALT HLA-DMB KCTD KIF NUP Calm1 (includes others) ADIPOR RNASE SGOL TRPM KIF18B SPAG Egln ARHGEF HDLBP TCN CDV NEK SOCS ABCB DNAJB MATK PPP1R15B ATL BIRC MYL CCNF MASTL SDCBP SELP DNAJC TRIM CELA KDELR RAPSN RTN ST3GAL PAFAH1B TGFB1 (includes EG:21803) RHOC Gm CCNO RAMP HK SPEF PQLC
41 SLC12A PYCR SH3GL DYNLT HSPA E2F EXOC TOB AHCY E2F RAB8B CTSB NOS INSL ZCCHC FUCA TCEB TRAPPC6A GFOD FKBP TMBIM MXI LAIR PHLPP IER SYK ADC DOK PGCP DNAJC Klra4 (includes others) MYO5A FAM72A LRP INHA CD TRIP PDK UAP DLGAP SMO UCK C1QTNF COX6B ITIH ZFAND OAS SAR1A SLC25A DNAJA ELOVL LSS IFIH
42 C11orf WSB KRTCAP SAMD CPEB LANCL SCARB EPS8L SLC7A IRAK HSPH ZFP SGMS RAB11FIP FAM213A GBE B9D MAD2L THOP MLEC PTGS BACH YKT STMN AP2B RAPGEF RAB11FIP NDFIP IMMT NUS UBE2A GPR137B SLC16A THEMIS MVP GOSR PLTP DUT SPHK PPP1R16B GLIS GSTO B01Rik LTBP SLC22A TMEM38B ACVRL CYB5B RAB7A SPSB TRAFD LTA SMPD
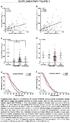
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Nature Medicine doi: /nm.2772
 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion
 Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
500 New CUSABIO Antibodies at MedStore!
 BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Supplementary Figure 1
 NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
Nature Immunology: doi: /ni Supplementary Figure 1. Foxm1 loss leads to bone marrow hypocellularity.
 Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplemental Information. A p53 Super-tumor Suppressor Reveals a Tumor. Suppressive p53-ptpn14-yap Axis. in Pancreatic Cancer
 Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
2 Composition. Invertible Mappings
 Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Figure 3 Three observations (Vp, Vs and density isosurfaces) intersecting in the PLF space. Solutions exist at the two indicated points.
 φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
ΠΕΡΙΕΧΟΜΕΝΑ. Κεφάλαιο 1: Κεφάλαιο 2: Κεφάλαιο 3:
 4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
[1] P Q. Fig. 3.1
![[1] P Q. Fig. 3.1 [1] P Q. Fig. 3.1](/thumbs/79/80362156.jpg) 1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
1 (a) Define resistance....... [1] (b) The smallest conductor within a computer processing chip can be represented as a rectangular block that is one atom high, four atoms wide and twenty atoms long. One
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ. Ψηφιακή Οικονομία. Διάλεξη 10η: Basics of Game Theory part 2 Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Ψηφιακή Οικονομία Διάλεξη 0η: Basics of Game Theory part 2 Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών Best Response Curves Used to solve for equilibria in games
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Ψηφιακή Οικονομία Διάλεξη 0η: Basics of Game Theory part 2 Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών Best Response Curves Used to solve for equilibria in games
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
Second Order RLC Filters
 ECEN 60 Circuits/Electronics Spring 007-0-07 P. Mathys Second Order RLC Filters RLC Lowpass Filter A passive RLC lowpass filter (LPF) circuit is shown in the following schematic. R L C v O (t) Using phasor
ECEN 60 Circuits/Electronics Spring 007-0-07 P. Mathys Second Order RLC Filters RLC Lowpass Filter A passive RLC lowpass filter (LPF) circuit is shown in the following schematic. R L C v O (t) Using phasor
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
The Simply Typed Lambda Calculus
 Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Supplementary Online Content
 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Table 1. Composition of the diets
 Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Materials for
 www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ
 ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
Prostate-targeted radiosensitization via aptamer-shrna chimeras in human. tumor xenografts
 SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
3.4 SUM AND DIFFERENCE FORMULAS. NOTE: cos(α+β) cos α + cos β cos(α-β) cos α -cos β
 3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
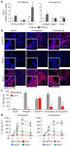 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1
 Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Zhang et al., Supplemental figures, Methods, References and Tables
 1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS
 CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
Supplementary Information Titles
 Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
ΚΥΠΡΙΑΚΟΣ ΣΥΝΔΕΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY 21 ος ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ Δεύτερος Γύρος - 30 Μαρτίου 2011
 Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Διάρκεια Διαγωνισμού: 3 ώρες Απαντήστε όλες τις ερωτήσεις Μέγιστο Βάρος (20 Μονάδες) Δίνεται ένα σύνολο από N σφαιρίδια τα οποία δεν έχουν όλα το ίδιο βάρος μεταξύ τους και ένα κουτί που αντέχει μέχρι
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3
 Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
Right Rear Door. Let's now finish the door hinge saga with the right rear door
 Right Rear Door Let's now finish the door hinge saga with the right rear door You may have been already guessed my steps, so there is not much to describe in detail. Old upper one file:///c /Documents
Right Rear Door Let's now finish the door hinge saga with the right rear door You may have been already guessed my steps, so there is not much to describe in detail. Old upper one file:///c /Documents
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Aluminum Electrolytic Capacitors (Large Can Type)
 Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Supplementary Figure 1.
 Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in. and *Christina Chan
 Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
Instruction Execution Times
 1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
Homework 3 Solutions
 Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Απόκριση σε Μοναδιαία Ωστική Δύναμη (Unit Impulse) Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο. Απόστολος Σ.
 Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο The time integral of a force is referred to as impulse, is determined by and is obtained from: Newton s 2 nd Law of motion states that the action
Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο The time integral of a force is referred to as impulse, is determined by and is obtained from: Newton s 2 nd Law of motion states that the action
Aluminum Electrolytic Capacitors
 Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Section 7.6 Double and Half Angle Formulas
 09 Section 7. Double and Half Angle Fmulas To derive the double-angles fmulas, we will use the sum of two angles fmulas that we developed in the last section. We will let α θ and β θ: cos(θ) cos(θ + θ)
09 Section 7. Double and Half Angle Fmulas To derive the double-angles fmulas, we will use the sum of two angles fmulas that we developed in the last section. We will let α θ and β θ: cos(θ) cos(θ + θ)
BD Cytometric Bead Array Flex Set System
 BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
6.1. Dirac Equation. Hamiltonian. Dirac Eq.
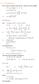 6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
Hydrogen Sorption Efficiency of Titanium Decorated Calix[4]pyrroles
![Hydrogen Sorption Efficiency of Titanium Decorated Calix[4]pyrroles Hydrogen Sorption Efficiency of Titanium Decorated Calix[4]pyrroles](/thumbs/84/89186739.jpg) Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 Hydrogen Sorption Efficiency of Titanium Decorated Calix[4]pyrroles Sandeep Kumar,
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 Hydrogen Sorption Efficiency of Titanium Decorated Calix[4]pyrroles Sandeep Kumar,
A Bonus-Malus System as a Markov Set-Chain. Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics
 A Bonus-Malus System as a Markov Set-Chain Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics Contents 1. Markov set-chain 2. Model of bonus-malus system 3. Example 4. Conclusions
A Bonus-Malus System as a Markov Set-Chain Małgorzata Niemiec Warsaw School of Economics Institute of Econometrics Contents 1. Markov set-chain 2. Model of bonus-malus system 3. Example 4. Conclusions
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
EE512: Error Control Coding
 EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
EE512: Error Control Coding Solution for Assignment on Finite Fields February 16, 2007 1. (a) Addition and Multiplication tables for GF (5) and GF (7) are shown in Tables 1 and 2. + 0 1 2 3 4 0 0 1 2 3
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine
 Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ. Ψηφιακή Οικονομία. Διάλεξη 7η: Consumer Behavior Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Ψηφιακή Οικονομία Διάλεξη 7η: Consumer Behavior Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών Τέλος Ενότητας Χρηματοδότηση Το παρόν εκπαιδευτικό υλικό έχει αναπτυχθεί
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Ψηφιακή Οικονομία Διάλεξη 7η: Consumer Behavior Mαρίνα Μπιτσάκη Τμήμα Επιστήμης Υπολογιστών Τέλος Ενότητας Χρηματοδότηση Το παρόν εκπαιδευτικό υλικό έχει αναπτυχθεί
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
(1) Describe the process by which mercury atoms become excited in a fluorescent tube (3)
 Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
Calculating the propagation delay of coaxial cable
 Your source for quality GNSS Networking Solutions and Design Services! Page 1 of 5 Calculating the propagation delay of coaxial cable The delay of a cable or velocity factor is determined by the dielectric
Your source for quality GNSS Networking Solutions and Design Services! Page 1 of 5 Calculating the propagation delay of coaxial cable The delay of a cable or velocity factor is determined by the dielectric
Reminders: linear functions
 Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
