Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells
|
|
|
- Ζαρά Σπανός
- 7 χρόνια πριν
- Προβολές:
Transcript
1 Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee A. 1,5, Gowda H. 1,5, Thiery JP. 6,7,8, Kumar P 1. 1 Institute of Bioinformatics, International Technology Park, Whitefield, Bangalore, , India. 2 Manipal University, Madhav Nagar, Manipal, , India. 3 Center for Bioinformatics, Pondicherry University, Puducherry , India. 4 Department of Molecular Reproduction, Development and Genetics, Indian Institute of Science, Bangalore, , India. 5 YU-IOB Center for Systems Biology and Molecular Medicine, Yenepoya University, Mangalore, , India. 6 Cancer Science Institute of Singapore, National University of Singapore, Centre for Translational Medicine NUS Yong Loo Lin School of Medicine, Singapore , Singapore. 7 Comprehensive Cancer Center, Institut Gustave Roussy, 114 Rue Edouard Vaillant, Villejuif, France; CNRS UMR 7057, Matter and Complex Systems, Université Paris Diderot, 10 rue Alice Domon et Léonie Duquet Paris, France. 8 Department of Biochemistry, National University of Singapore, Singapore , Singapore. Correspondence and requests for materials should be addressed to Kumar P. ( prashant@ibioinformatics.org) 1
2 Supplementary Information As shown in the supplementary figure 1a, the total number of papers that resulted from PubMed search with keyword circulating tumor cells signaling is 1,073 (blue portion of the Venn diagram; publications). This includes the papers that were common to results from keyword searches circulating tumor cells signaling and cancer (648); circulating tumor cells signaling and epithelial mesenchymal transition (2); and results common to all three keyword searches (45). The papers unique to the last two searches were excluded as they were not relevant. After abstract-level screening of these 1,073 papers, 153 were narrowed down for in-depth screening. Similarly, as shown in the supplementary figure 1b, the total number of papers that resulted from PubMed search with keyword circulating tumor cells gene expression is 2,169 (blue portion of the Venn diagram; ,043 publications). This includes the papers that were common to results from keyword searches circulating tumor cells genes (214); circulating tumor cells expression (1,043); and results common to all three keyword searches (424). The papers unique to the last two searches were excluded due to non-relevance and large numbers. After abstract-level screening of these 2,169 papers, 78 were narrowed down for in-depth screening. Out of these 231 papers (153+78), 98 papers were found to have gene expression data relevant to CTCs from which the gene list was compiled for our analysis. Of these, 93 papers were smallscale qualitative and semi-quantitative experiments that reported expression of a few genes in CTCs. The remaining five papers were based on large-scale transcriptomic experiments (RNA- Seq and Microarray) and reported differential expression of hundreds of genes in CTCs. The final gene list for our pathway analysis comprises mostly of genes from these five papers (7,209 out 7,572 total numbers of genes). These numbers are reported as CTC_ALL (7,572 genes from all 98 papers) out of which 7,209 (CTC_FC) were taken from the five large-scale transcriptomics experiment papers (Supplementary Table S1 and Supplementary Table S2, respectively). The reference information (PubMed IDs; PMIDs) for the 98 papers are listed in Supplementary Table S5. 2
3 Supplementary Figures Supplementary Figure 1: Venn diagrams showing overlap between research articles when searched with keywords in Pubmed. (a) PubMed results with keywords containing circulating tumor cells signaling ; (b) PubMed results with keywords containing circulating tumor cells gene expression ; (c) Venn diagram showing number of differentially expressed genes in circulating tumor cells taken from semi-quantitative and qualitative experiments shown in CTC_ALL (n=7,572). Subset of differentially expressed genes from semi-quantitative experiments from five cancer studies is shown as CTC_FC (n=7,209). 3
4 Supplementary Figure 2: Venn diagram showing the number of genes differentially expressed in CTCs in the five cancers. Number of genes overexpressed in CTCs when compared to primary tumors (a), peripheral blood mononuclear cells (PBMCs) (b), normal tissue (c). Number of genes downregulated in CTCs when compared to primary tumors (d), PBMCs (e), normal tissue (f). 4
5 Supplementary Figure 3: Swarm plot shows the distribution of differentially expressed epithelial (green) and mesenchymal (blue) genes across the five cancer datasets. The genes are further segregated on their expression as epithelial or mesenchymal markers based on published literature. PRCTC1 denotes prostate CTCs gene expression when compared to primary tumors; CRCTC1 colorectal CTCs when compared to PBMCs; MNCTC1 melanoma CTCs/melanoma cell lines; MNCTC2 melanoma CTCs/primary melanocytes; PACTC1 pancreatic CTCs/nontumoral pancreatic tissue; PACTC2 pancreatic CTCs/PBMCs; PACTC3 pancreatic CTCs/primary tumors; BRCTC1 breast CTCs/breast epithelium; BRCTC2 breast CTCs/PBMCs; BRCTC3 breast CTCs/primary tumors. 5
6 Supplementary Figure 4: Comparison of EMT scores of CTC gene expression data for the five cancers with a) EMT scores of tumors and b) EMT scores of cancer cell lines. The CTC dataset labels are highlighted in boxes. 6
7 Supplementary Tables Supplementary Table S1: CTC_ALL: Differentially expressed genes in circulating tumor cells when compared with primary tumors, PBMCs, normal tissue and tumor cell lines. Expression data for a total number of 7,572 genes were obtained from reported literature of semiquantitative and qualitative experiments. Supplementary Table S2: CTC_FC: Differentially expressed genes in circulating tumor cells when compared with primary tumors, PBMCs, normal tissue and tumor cell lines. Data for a total of 7,209 genes were obtained from reported literature of transcriptomics experiments from five different cancer studies of CTCs. Supplementary Table S3: CTC isolation and purity confirmation techniques used by the five high-throughput experiments from which differentially expressed genes with fold change values (CTC_FC) was compiled for data analysis in our study. Supplementary Table S4: List of molecular interactions reported in the literature (PubMed IDs included) and depicted in the leukocyte/ctc extravasation pathway. Supplementary Table S5: PubMed IDs for 98 papers from which a gene list was compiled for molecular characterization of CTCs. This list includes the five large-scale transcriptomics experiment papers from which differentially expressed genes with fold change values were obtained (highlighted in yellow color). Supplementary Table S6: EMT scores computed from transcriptome datasets of CTCs in five cancers GSE18670, GSE31023, GSE38495, GSE45965, and GSE The pre-processed data GSE45965 was also used. 7
8 Yadavalli et al., Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Supplementary Table S1: CTC_ALL: Differentially expressed genes in circulating tumor cells when compared to primary tumors, PBMC, normal tissue and tumor cell lines. Expression data for a total number of 7,572 genes were obtained from reported literature of semi-quantitative and qualitative experiments. Gene Symbol Gene ID Gene Description Fold change (log2) Arbitrary fold change (for CTC_ALL analysis) 1 RPS4Y ribosomal protein S4, Y-linked 1 [Source:HGNC Symbol;Acc:HGNC:10425] COX7B cytochrome c oxidase subunit 7B2 [Source:HGNC Symbol;Acc:HGNC:24381] GTSF gametocyte specific factor 1 [Source:HGNC Symbol;Acc:HGNC:26565] SMUG single-strand-selective monofunctional uracil-dna glycosylase 1 [Source:HGNC Symbol;Acc:HGNC:17148] PAGE PAGE family member 5 [Source:HGNC Symbol;Acc:HGNC:29992] TUBB tubulin beta 1 class VI [Source:HGNC Symbol;Acc:HGNC:16257] PPBP 5473 pro-platelet basic protein [Source:HGNC Symbol;Acc:HGNC:9240] TMEFF transmembrane protein with EGF like and two follistatin like domains 2 [Source:HGNC Symbol;Acc:HGNC:11867] PF platelet factor 4 [Source:HGNC Symbol;Acc:HGNC:8861] ANKHD ankyrin repeat and KH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:24714] ANKHD1-EIF4EBP ANKHD1-EIF4EBP3 readthrough [Source:HGNC Symbol;Acc:HGNC:33530] MLEC 9761 malectin [Source:HGNC Symbol;Acc:HGNC:28973] S100A S100 calcium binding protein A12 [Source:HGNC Symbol;Acc:HGNC:10489] FCN ficolin 1 [Source:HGNC Symbol;Acc:HGNC:3623] ACRBP acrosin binding protein [Source:HGNC Symbol;Acc:HGNC:17195] CCNB1 891 cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] NRGN 4900 neurogranin [Source:HGNC Symbol;Acc:HGNC:8000] SREK splicing regulatory glutamic acid and lysine rich protein 1 [Source:HGNC Symbol;Acc:HGNC:17882] CENPM centromere protein M [Source:HGNC Symbol;Acc:HGNC:18352] ANLN anillin actin binding protein [Source:HGNC Symbol;Acc:HGNC:14082] KIAA KIAA0895 [Source:HGNC Symbol;Acc:HGNC:22206] LYZ 4069 lysozyme [Source:HGNC Symbol;Acc:HGNC:6740] CKAP cytoskeleton-associated protein 4 [Source:HGNC Symbol;Acc:HGNC:16991] ITGA2B 3674 integrin subunit alpha 2b [Source:HGNC Symbol;Acc:HGNC:6138] TSG tumor susceptibility 101 [Source:HGNC Symbol;Acc:HGNC:15971] SRSF serine and arginine rich splicing factor 11 [Source:HGNC Symbol;Acc:HGNC:10782] SNX sorting nexin 10 [Source:HGNC Symbol;Acc:HGNC:14974] BMX 660 BMX non-receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:1079] HIST1H3H 8357 histone cluster 1, H3h [Source:HGNC Symbol;Acc:HGNC:4775] ECI enoyl-coa delta isomerase 2 [Source:HGNC Symbol;Acc:HGNC:14601] RUVBL RuvB like AAA ATPase 1 [Source:HGNC Symbol;Acc:HGNC:10474] MRPL mitochondrial ribosomal protein L46 [Source:HGNC Symbol;Acc:HGNC:1192] MAF MAF1 homolog, negative regulator of RNA polymerase III [Source:HGNC Symbol;Acc:HGNC:24966] PF4V platelet factor 4 variant 1 [Source:HGNC Symbol;Acc:HGNC:8862] YY1AP YY1 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:30935] TIMMDC translocase of inner mitochondrial membrane domain containing 1 [Source:HGNC Symbol;Acc:HGNC:1321] LYAR Ly1 antibody reactive [Source:HGNC Symbol;Acc:HGNC:26021] TAL T-cell acute lymphocytic leukemia 1 [Source:HGNC Symbol;Acc:HGNC:11556] SF3B splicing factor 3b subunit 6 [Source:HGNC Symbol;Acc:HGNC:30096] BUD BUD31 homolog [Source:HGNC Symbol;Acc:HGNC:29629] RPL10L ribosomal protein L10 like [Source:HGNC Symbol;Acc:HGNC:17976] MGAM 8972 maltase-glucoamylase [Source:HGNC Symbol;Acc:HGNC:7043] ALOX arachidonate 12-lipoxygenase, 12S type [Source:HGNC Symbol;Acc:HGNC:429] CCNB cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] CYFIP cytoplasmic FMR1 interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:13760] TREML triggering receptor expressed on myeloid cells like 1 [Source:HGNC Symbol;Acc:HGNC:20434] DNAJA DnaJ heat shock protein family (Hsp40) member A4 [Source:HGNC Symbol;Acc:HGNC:14885] HBG hemoglobin subunit gamma 1 [Source:HGNC Symbol;Acc:HGNC:4831] FUNDC FUN14 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:24925] RGS regulator of G-protein signaling 18 [Source:HGNC Symbol;Acc:HGNC:14261] KRTAP keratin associated protein 19-1 [Source:HGNC Symbol;Acc:HGNC:18936] LPXN 9404 leupaxin [Source:HGNC Symbol;Acc:HGNC:14061] SLC35B solute carrier family 35 member B1 [Source:HGNC Symbol;Acc:HGNC:20798] USP ubiquitin specific peptidase 15 [Source:HGNC Symbol;Acc:HGNC:12613] NAMPT nicotinamide phosphoribosyltransferase [Source:HGNC Symbol;Acc:HGNC:30092] AHSP alpha hemoglobin stabilizing protein [Source:HGNC Symbol;Acc:HGNC:18075] EWSR EWS RNA binding protein 1 [Source:HGNC Symbol;Acc:HGNC:3508] CELF CUGBP, Elav-like family member 1 [Source:HGNC Symbol;Acc:HGNC:2549] IFT intraflagellar transport 57 [Source:HGNC Symbol;Acc:HGNC:17367] SLA Src-like-adaptor 2 [Source:HGNC Symbol;Acc:HGNC:17329] SERPINA3 12 serpin family A member 3 [Source:HGNC Symbol;Acc:HGNC:16] HMGN high mobility group nucleosomal binding domain 4 [Source:HGNC Symbol;Acc:HGNC:4989] TPP tripeptidyl peptidase 2 [Source:HGNC Symbol;Acc:HGNC:12016] NLRC NLR family CARD domain containing 5 [Source:HGNC Symbol;Acc:HGNC:29933] GNAZ 2781 G protein subunit alpha z [Source:HGNC Symbol;Acc:HGNC:4395] SERPINB serpin family B member 6 [Source:HGNC Symbol;Acc:HGNC:8950] CDKN cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] PNN 5411 pinin, desmosome associated protein [Source:HGNC Symbol;Acc:HGNC:9162] TSTD thiosulfate sulfurtransferase like domain containing 1 [Source:HGNC Symbol;Acc:HGNC:35410] CST cystatin F [Source:HGNC Symbol;Acc:HGNC:2479] 5.4 2
9 71 IGF2BP insulin like growth factor 2 mrna binding protein 3 [Source:HGNC Symbol;Acc:HGNC:28868] CIAPIN cytokine induced apoptosis inhibitor 1 [Source:HGNC Symbol;Acc:HGNC:28050] RAB RAB37, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:30268] SDPR 8436 serum deprivation response [Source:HGNC Symbol;Acc:HGNC:10690] AP3D adaptor related protein complex 3 delta 1 subunit [Source:HGNC Symbol;Acc:HGNC:568] MSL male specific lethal 1 homolog [Source:HGNC Symbol;Acc:HGNC:27905] PSMB proteasome subunit beta 8 [Source:HGNC Symbol;Acc:HGNC:9545] HIST1H2AG 8969 histone cluster 1, H2ag [Source:HGNC Symbol;Acc:HGNC:4737] HMMR 3161 hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] IL2RB 3560 interleukin 2 receptor subunit beta [Source:HGNC Symbol;Acc:HGNC:6009] RBM RNA binding motif protein 18 [Source:HGNC Symbol;Acc:HGNC:28413] BUB1 699 BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] MAGEA MAGE family member A10 [Source:HGNC Symbol;Acc:HGNC:6797] MAGEA10-MAGEA MAGEA10-MAGEA5 Readthrough ANKRD ankyrin repeat domain 17 [Source:HGNC Symbol;Acc:HGNC:23575] TMEM transmembrane protein 40 [Source:HGNC Symbol;Acc:HGNC:25620] AFTPH aftiphilin [Source:HGNC Symbol;Acc:HGNC:25951] ATP2B1 490 ATPase plasma membrane Ca2+ transporting 1 [Source:HGNC Symbol;Acc:HGNC:814] DCAF DDB1 and CUL4 associated factor 13 [Source:HGNC Symbol;Acc:HGNC:24535] SLC25A solute carrier family 25 member 32 [Source:HGNC Symbol;Acc:HGNC:29683] GFI1B 8328 growth factor independent 1B transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:4238] S100A S100 calcium binding protein A8 [Source:HGNC Symbol;Acc:HGNC:10498] TMEM87A transmembrane protein 87A [Source:HGNC Symbol;Acc:HGNC:24522] ANKRD ankyrin repeat domain 10 [Source:HGNC Symbol;Acc:HGNC:20265] PTN 5764 pleiotrophin [Source:HGNC Symbol;Acc:HGNC:9630] TRIP thyroid hormone receptor interactor 12 [Source:HGNC Symbol;Acc:HGNC:12306] KRT keratin 15 [Source:HGNC Symbol;Acc:HGNC:6421] ITGB integrin subunit beta 2 [Source:HGNC Symbol;Acc:HGNC:6155] WDR45B WD repeat domain 45B [Source:HGNC Symbol;Acc:HGNC:25072] IFT intraflagellar transport 20 [Source:HGNC Symbol;Acc:HGNC:30989] FGR 2268 FGR proto-oncogene, Src family tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:3697] NR2C nuclear receptor subfamily 2 group C member 2 [Source:HGNC Symbol;Acc:HGNC:7972] TRA2A transformer 2 alpha homolog [Source:HGNC Symbol;Acc:HGNC:16645] MT-ND mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 [Source:HGNC Symbol;Acc:HGNC:7462] NUP nucleoporin 93 [Source:HGNC Symbol;Acc:HGNC:28958] MAGEB MAGE family member B1 [Source:HGNC Symbol;Acc:HGNC:6808] MAGEB MAGE family member B4 [Source:HGNC Symbol;Acc:HGNC:6811] ATP5S ATP synthase, H+ transporting, mitochondrial Fo complex subunit s (factor B) [Source:HGNC Symbol;Acc:HGNC:18799] C2orf chromosome 2 open reading frame 88 [Source:HGNC Symbol;Acc:HGNC:28191] PAEP 5047 progestagen associated endometrial protein [Source:HGNC Symbol;Acc:HGNC:8573] SAMD sterile alpha motif domain containing 14 [Source:HGNC Symbol;Acc:HGNC:27312] TMEM106C transmembrane protein 106C [Source:HGNC Symbol;Acc:HGNC:28775] SPPL2B signal peptide peptidase like 2B [Source:HGNC Symbol;Acc:HGNC:30627] CD CD247 molecule [Source:HGNC Symbol;Acc:HGNC:1677] NUSAP nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] KIF5C 3800 kinesin family member 5C [Source:HGNC Symbol;Acc:HGNC:6325] TRIM tripartite motif containing 58 [Source:HGNC Symbol;Acc:HGNC:24150] MKKS 8195 McKusick-Kaufman syndrome [Source:HGNC Symbol;Acc:HGNC:7108] ERN endoplasmic reticulum to nucleus signaling 2 [Source:HGNC Symbol;Acc:HGNC:16942] PLK polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] POM POM121 transmembrane nucleoporin [Source:HGNC Symbol;Acc:HGNC:19702] TMEM transmembrane protein 11 [Source:HGNC Symbol;Acc:HGNC:16823] SNRNP small nuclear ribonucleoprotein U11/U12 subunit 25 [Source:HGNC Symbol;Acc:HGNC:14161] PRAM PML-RARA regulated adaptor molecule 1 [Source:HGNC Symbol;Acc:HGNC:30091] GMPR guanosine monophosphate reductase 2 [Source:HGNC Symbol;Acc:HGNC:4377] ILF interleukin enhancer binding factor 3 [Source:HGNC Symbol;Acc:HGNC:6038] LUZP leucine zipper protein 4 [Source:HGNC Symbol;Acc:HGNC:24971] NUP nucleoporin 35 [Source:HGNC Symbol;Acc:HGNC:29797] SMPD sphingomyelin phosphodiesterase 1 [Source:HGNC Symbol;Acc:HGNC:11120] MBD methyl-cpg binding domain protein 6 [Source:HGNC Symbol;Acc:HGNC:20445] APOBEC3A apolipoprotein B mrna editing enzyme catalytic subunit 3A [Source:HGNC Symbol;Acc:HGNC:17343] UNC13B unc-13 homolog B (C. elegans) [Source:HGNC Symbol;Acc:HGNC:12566] TIMM8B translocase of inner mitochondrial membrane 8 homolog B (yeast) [Source:HGNC Symbol;Acc:HGNC:11818] C6orf chromosome 6 open reading frame 62 [Source:HGNC Symbol;Acc:HGNC:20998] YWHAG 7532 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma [Source:HGNC Symbol;Acc:HGNC:12852] FAM133B family with sequence similarity 133 member B [Source:HGNC Symbol;Acc:HGNC:28629] CKAP cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] MAGEC MAGE family member C2 [Source:HGNC Symbol;Acc:HGNC:13574] ARL6IP ADP ribosylation factor like GTPase 6 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:697] ALDH9A1 223 aldehyde dehydrogenase 9 family member A1 [Source:HGNC Symbol;Acc:HGNC:412] UBE2V ubiquitin conjugating enzyme E2 V2 [Source:HGNC Symbol;Acc:HGNC:12495] LILRA leukocyte immunoglobulin like receptor A2 [Source:HGNC Symbol;Acc:HGNC:6603] GP glycoprotein IX platelet [Source:HGNC Symbol;Acc:HGNC:4444] PDZK1IP PDZK1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:16887] 4.9 2
10 145 MAGEB MAGE family member B2 [Source:HGNC Symbol;Acc:HGNC:6809] PPP2R2A 5520 protein phosphatase 2 regulatory subunit Balpha [Source:HGNC Symbol;Acc:HGNC:9304] C12orf chromosome 12 open reading frame 75 [Source:HGNC Symbol;Acc:HGNC:35164] PRKAR2B 5577 protein kinase camp-dependent type II regulatory subunit beta [Source:HGNC Symbol;Acc:HGNC:9392] CFP 5199 complement factor properdin [Source:HGNC Symbol;Acc:HGNC:8864] DNAJB DnaJ heat shock protein family (Hsp40) member B7 [Source:HGNC Symbol;Acc:HGNC:24986] XPNPEP X-prolyl aminopeptidase 3, mitochondrial [Source:HGNC Symbol;Acc:HGNC:28052] SELENBP selenium binding protein 1 [Source:HGNC Symbol;Acc:HGNC:10719] SMU DNA replication regulator and spliceosomal factor [Source:HGNC Symbol;Acc:HGNC:18247] SUMO small ubiquitin-like modifier 2 [Source:HGNC Symbol;Acc:HGNC:11125] SEC61B Sec61 translocon beta subunit [Source:HGNC Symbol;Acc:HGNC:16993] PEF penta-ef-hand domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30009] PRKCB 5579 protein kinase C beta [Source:HGNC Symbol;Acc:HGNC:9395] TOP2B 7155 topoisomerase (DNA) II beta [Source:HGNC Symbol;Acc:HGNC:11990] HK hexokinase 3 [Source:HGNC Symbol;Acc:HGNC:4925] SFSWAP 6433 splicing factor SWAP homolog [Source:HGNC Symbol;Acc:HGNC:10790] PRRC2C proline rich coiled-coil 2C [Source:HGNC Symbol;Acc:HGNC:24903] COA cytochrome c oxidase assembly factor 1 homolog [Source:HGNC Symbol;Acc:HGNC:21868] LILRB leukocyte immunoglobulin like receptor B2 [Source:HGNC Symbol;Acc:HGNC:6606] CCT chaperonin containing TCP1 subunit 2 [Source:HGNC Symbol;Acc:HGNC:1615] MAGED MAGE family member D2 [Source:HGNC Symbol;Acc:HGNC:16353] CDC cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] SNRPA small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] MAP3K7CL MAP3K7 C-terminal like [Source:HGNC Symbol;Acc:HGNC:16457] PADI peptidyl arginine deiminase 4 [Source:HGNC Symbol;Acc:HGNC:18368] SPX spexin hormone [Source:HGNC Symbol;Acc:HGNC:28139] GZMB 3002 granzyme B [Source:HGNC Symbol;Acc:HGNC:4709] RASGRP RAS guanyl releasing protein 2 [Source:HGNC Symbol;Acc:HGNC:9879] FAM133A family with sequence similarity 133 member A [Source:HGNC Symbol;Acc:HGNC:26748] MPL 4352 MPL proto-oncogene, thrombopoietin receptor [Source:HGNC Symbol;Acc:HGNC:7217] APOBEC3B 9582 apolipoprotein B mrna editing enzyme catalytic subunit 3B [Source:HGNC Symbol;Acc:HGNC:17352] PTGS prostaglandin-endoperoxide synthase 1 [Source:HGNC Symbol;Acc:HGNC:9604] TIMM17A translocase of inner mitochondrial membrane 17 homolog A (yeast) [Source:HGNC Symbol;Acc:HGNC:17315] NFE nuclear factor, erythroid 2 [Source:HGNC Symbol;Acc:HGNC:7780] MSN 4478 moesin [Source:HGNC Symbol;Acc:HGNC:7373] CCNL cyclin L1 [Source:HGNC Symbol;Acc:HGNC:20569] NUP nucleoporin 37 [Source:HGNC Symbol;Acc:HGNC:29929] TMEM transmembrane protein 91 [Source:HGNC Symbol;Acc:HGNC:32393] ZNF zinc finger protein 185 (LIM domain) [Source:HGNC Symbol;Acc:HGNC:12976] PMM phosphomannomutase 1 [Source:HGNC Symbol;Acc:HGNC:9114] USP ubiquitin specific peptidase 1 [Source:HGNC Symbol;Acc:HGNC:12607] DLGAP DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] TROAP trophinin associated protein [Source:HGNC Symbol;Acc:HGNC:12327] LUC7L LUC7 like 3 pre-mrna splicing factor [Source:HGNC Symbol;Acc:HGNC:24309] SRPRB SRP receptor beta subunit [Source:HGNC Symbol;Acc:HGNC:24085] NFIL nuclear factor, interleukin 3 regulated [Source:HGNC Symbol;Acc:HGNC:7787] ADD3 120 adducin 3 [Source:HGNC Symbol;Acc:HGNC:245] TAP transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) [Source:HGNC Symbol;Acc:HGNC:43] DLD 1738 dihydrolipoamide dehydrogenase [Source:HGNC Symbol;Acc:HGNC:2898] SLC25A solute carrier family 25 member 38 [Source:HGNC Symbol;Acc:HGNC:26054] PLOD procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 [Source:HGNC Symbol;Acc:HGNC:9081] PABPN poly(a) binding protein nuclear 1 [Source:HGNC Symbol;Acc:HGNC:8565] GAL3ST galactose-3-o-sulfotransferase 4 [Source:HGNC Symbol;Acc:HGNC:24145] DZIP DAZ interacting zinc finger protein 3 [Source:HGNC Symbol;Acc:HGNC:30938] KIAA KIAA1524 [Source:HGNC Symbol;Acc:HGNC:29302] C10orf chromosome 10 open reading frame 54 [Source:HGNC Symbol;Acc:HGNC:30085] MFAP3L 9848 microfibrillar associated protein 3 like [Source:HGNC Symbol;Acc:HGNC:29083] CTTN 2017 cortactin [Source:HGNC Symbol;Acc:HGNC:3338] VPS VPS28, ESCRT-I subunit [Source:HGNC Symbol;Acc:HGNC:18178] DCTN dynactin subunit 3 [Source:HGNC Symbol;Acc:HGNC:2713] CCDC coiled-coil domain containing 152 [Source:HGNC Symbol;Acc:HGNC:34438] SEPP selenoprotein P, plasma, 1 [Source:HGNC Symbol;Acc:HGNC:10751] PSMC proteasome 26S subunit, ATPase 2 [Source:HGNC Symbol;Acc:HGNC:9548] TMED transmembrane p24 trafficking protein 2 [Source:HGNC Symbol;Acc:HGNC:16996] EIF2S eukaryotic translation initiation factor 2 subunit beta [Source:HGNC Symbol;Acc:HGNC:3266] S100A S100 calcium binding protein A9 [Source:HGNC Symbol;Acc:HGNC:10499] SYTL synaptotagmin like 5 [Source:HGNC Symbol;Acc:HGNC:15589] RIN Ras and Rab interactor 3 [Source:HGNC Symbol;Acc:HGNC:18751] HNRNPH heterogeneous nuclear ribonucleoprotein H3 [Source:HGNC Symbol;Acc:HGNC:5043] CMTM CKLF like MARVEL transmembrane domain containing 5 [Source:HGNC Symbol;Acc:HGNC:19176] FAM136A family with sequence similarity 136 member A [Source:HGNC Symbol;Acc:HGNC:25911] SYCP synaptonemal complex protein 3 [Source:HGNC Symbol;Acc:HGNC:18130] C11orf chromosome 11 open reading frame 21 [Source:HGNC Symbol;Acc:HGNC:13231] GNG G protein subunit gamma 10 [Source:HGNC Symbol;Acc:HGNC:4402] 4.6 2
11 219 DNAJC25-GNG DNAJC25-GNG10 readthrough [Source:HGNC Symbol;Acc:HGNC:37501] DAPP dual adaptor of phosphotyrosine and 3-phosphoinositides 1 [Source:HGNC Symbol;Acc:HGNC:16500] MRPL mitochondrial ribosomal protein L22 [Source:HGNC Symbol;Acc:HGNC:14480] HJURP Holliday junction recognition protein [Source:HGNC Symbol;Acc:HGNC:25444] ALDH3A2 224 aldehyde dehydrogenase 3 family member A2 [Source:HGNC Symbol;Acc:HGNC:403] SPATA spermatogenesis associated 20 [Source:HGNC Symbol;Acc:HGNC:26125] C4orf chromosome 4 open reading frame 19 [Source:HGNC Symbol;Acc:HGNC:25618] RELL RELT like 1 [Source:HGNC Symbol;Acc:HGNC:27379] TSPAN tetraspanin 33 [Source:HGNC Symbol;Acc:HGNC:28743] CLEC12A C-type lectin domain family 12 member A [Source:HGNC Symbol;Acc:HGNC:31713] C19orf chromosome 19 open reading frame 33 [Source:HGNC Symbol;Acc:HGNC:16668] UGP UDP-glucose pyrophosphorylase 2 [Source:HGNC Symbol;Acc:HGNC:12527] CCDC coiled-coil domain containing 169 [Source:HGNC Symbol;Acc:HGNC:34361] CCDC169-SOHLH CCDC169-SOHLH2 readthrough [Source:HGNC Symbol;Acc:HGNC:38866] SOHLH spermatogenesis and oogenesis specific basic helix-loop-helix 2 [Source:HGNC Symbol;Acc:HGNC:26026] NOV 4856 nephroblastoma overexpressed [Source:HGNC Symbol;Acc:HGNC:7885] ATF3 467 activating transcription factor 3 [Source:HGNC Symbol;Acc:HGNC:785] PSMA proteasome subunit alpha 5 [Source:HGNC Symbol;Acc:HGNC:9534] AURKA 6790 aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] PRF perforin 1 [Source:HGNC Symbol;Acc:HGNC:9360] DNAJA DnaJ heat shock protein family (Hsp40) member A2 [Source:HGNC Symbol;Acc:HGNC:14884] CSF3R 1441 colony stimulating factor 3 receptor [Source:HGNC Symbol;Acc:HGNC:2439] CHORDC cysteine and histidine rich domain containing 1 [Source:HGNC Symbol;Acc:HGNC:14525] NCF neutrophil cytosolic factor 4 [Source:HGNC Symbol;Acc:HGNC:7662] RACGAP Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] TCEAL transcription elongation factor A like 8 [Source:HGNC Symbol;Acc:HGNC:28683] S100A S100 calcium binding protein A16 [Source:HGNC Symbol;Acc:HGNC:20441] GFOD glucose-fructose oxidoreductase domain containing 1 [Source:HGNC Symbol;Acc:HGNC:21096] RUFY RUN and FYVE domain containing 1 [Source:HGNC Symbol;Acc:HGNC:19760] EIF eukaryotic translation initiation factor 5 [Source:HGNC Symbol;Acc:HGNC:3299] PGM phosphoglucomutase 2 [Source:HGNC Symbol;Acc:HGNC:8906] ASNS 440 asparagine synthetase (glutamine-hydrolyzing) [Source:HGNC Symbol;Acc:HGNC:753] LILRA leukocyte immunoglobulin like receptor A3 [Source:HGNC Symbol;Acc:HGNC:6604] MRRF mitochondrial ribosome recycling factor [Source:HGNC Symbol;Acc:HGNC:7234] YY YY1 transcription factor [Source:HGNC Symbol;Acc:HGNC:12856] TOP topoisomerase (DNA) I [Source:HGNC Symbol;Acc:HGNC:11986] FAM192A family with sequence similarity 192 member A [Source:HGNC Symbol;Acc:HGNC:29856] DRAP DR1 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:3019] OGT 8473 O-linked N-acetylglucosamine (GlcNAc) transferase [Source:HGNC Symbol;Acc:HGNC:8127] TCAF TRPM8 channel associated factor 1 [Source:HGNC Symbol;Acc:HGNC:22201] ITM2A 9452 integral membrane protein 2A [Source:HGNC Symbol;Acc:HGNC:6173] SLC25A solute carrier family 25 member 36 [Source:HGNC Symbol;Acc:HGNC:25554] RAP1GDS Rap1 GTPase-GDP dissociation stimulator 1 [Source:HGNC Symbol;Acc:HGNC:9859] PPM1A 5494 protein phosphatase, Mg2+/Mn2+ dependent 1A [Source:HGNC Symbol;Acc:HGNC:9275] DPEP dipeptidase 2 [Source:HGNC Symbol;Acc:HGNC:23028] HBA hemoglobin subunit alpha 2 [Source:HGNC Symbol;Acc:HGNC:4824] UBE2E ubiquitin conjugating enzyme E2 E3 [Source:HGNC Symbol;Acc:HGNC:12479] WIPI WD repeat domain, phosphoinositide interacting 1 [Source:HGNC Symbol;Acc:HGNC:25471] CHD chromodomain helicase DNA binding protein 4 [Source:HGNC Symbol;Acc:HGNC:1919] GP1BA 2811 glycoprotein Ib platelet alpha subunit [Source:HGNC Symbol;Acc:HGNC:4439] BEND BEN domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28509] COMMD COMM domain containing 7 [Source:HGNC Symbol;Acc:HGNC:16223] FLII 2314 FLII, actin remodeling protein [Source:HGNC Symbol;Acc:HGNC:3750] CHMP charged multivesicular body protein 5 [Source:HGNC Symbol;Acc:HGNC:26942] ADRM adhesion regulating molecule 1 [Source:HGNC Symbol;Acc:HGNC:15759] GZMH 2999 granzyme H [Source:HGNC Symbol;Acc:HGNC:4710] RABIF 5877 RAB interacting factor [Source:HGNC Symbol;Acc:HGNC:9797] PBK PDZ binding kinase [Source:HGNC Symbol;Acc:HGNC:18282] PROK prokineticin 2 [Source:HGNC Symbol;Acc:HGNC:18455] ANKRD ankyrin repeat domain 55 [Source:HGNC Symbol;Acc:HGNC:25681] PAGE2B PAGE family member 2B [Source:HGNC Symbol;Acc:HGNC:31805] GNLY granulysin [Source:HGNC Symbol;Acc:HGNC:4414] SSB 6741 Sjogren syndrome antigen B [Source:HGNC Symbol;Acc:HGNC:11316] RBM RNA binding motif protein 22 [Source:HGNC Symbol;Acc:HGNC:25503] MCL myeloid cell leukemia 1 [Source:HGNC Symbol;Acc:HGNC:6943] OSTC oligosaccharyltransferase complex subunit (non-catalytic) [Source:HGNC Symbol;Acc:HGNC:24448] PILRB paired immunoglobin-like type 2 receptor beta [Source:HGNC Symbol;Acc:HGNC:18297] SPDYE speedy/ringo cell cycle regulator family member E3 [Source:HGNC Symbol;Acc:HGNC:35462] ZNF zinc finger protein 467 [Source:HGNC Symbol;Acc:HGNC:23154] SRPX 8406 sushi repeat containing protein, X-linked [Source:HGNC Symbol;Acc:HGNC:11309] CCDC coiled-coil domain containing 73 [Source:HGNC Symbol;Acc:HGNC:23261] TTK 7272 TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] AKIRIN akirin 1 [Source:HGNC Symbol;Acc:HGNC:25744] IST IST1, ESCRT-III associated factor [Source:HGNC Symbol;Acc:HGNC:28977] SRP signal recognition particle 72kDa [Source:HGNC Symbol;Acc:HGNC:11303] CLUL clusterin like 1 [Source:HGNC Symbol;Acc:HGNC:2096] 4.3 2
12 295 ENOSF enolase superfamily member 1 [Source:HGNC Symbol;Acc:HGNC:30365] TYMS 7298 thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] TYMSOS TYMS opposite strand [Source:HGNC Symbol;Acc:HGNC:29553] APOD 347 apolipoprotein D [Source:HGNC Symbol;Acc:HGNC:612] CENPK centromere protein K [Source:HGNC Symbol;Acc:HGNC:29479] SURF surfeit 2 [Source:HGNC Symbol;Acc:HGNC:11475] C5orf chromosome 5 open reading frame 15 [Source:HGNC Symbol;Acc:HGNC:20656] LARP La ribonucleoprotein domain family member 4 [Source:HGNC Symbol;Acc:HGNC:24320] FRMD FERM domain containing 3 [Source:HGNC Symbol;Acc:HGNC:24125] CTNNAL catenin alpha like 1 [Source:HGNC Symbol;Acc:HGNC:2512] LGMN 5641 legumain [Source:HGNC Symbol;Acc:HGNC:9472] MMP matrix metallopeptidase 9 [Source:HGNC Symbol;Acc:HGNC:7176] C8orf chromosome 8 open reading frame 76 [Source:HGNC Symbol;Acc:HGNC:25924] ZHX zinc fingers and homeoboxes 1 [Source:HGNC Symbol;Acc:HGNC:12871] ZHX1-C8orf ZHX1-C8orf76 readthrough [Source:HGNC Symbol;Acc:HGNC:42975] HN hematological and neurological expressed 1 [Source:HGNC Symbol;Acc:HGNC:14569] TNFRSF1B 7133 tumor necrosis factor receptor superfamily member 1B [Source:HGNC Symbol;Acc:HGNC:11917] MSMO methylsterol monooxygenase 1 [Source:HGNC Symbol;Acc:HGNC:10545] ARL4C ADP ribosylation factor like GTPase 4C [Source:HGNC Symbol;Acc:HGNC:698] LTB4R 1241 leukotriene B4 receptor [Source:HGNC Symbol;Acc:HGNC:6713] TTC tetratricopeptide repeat domain 1 [Source:HGNC Symbol;Acc:HGNC:12391] HMGB high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] TMEM170B transmembrane protein 170B [Source:HGNC Symbol;Acc:HGNC:34244] PUM pumilio RNA binding family member 3 [Source:HGNC Symbol;Acc:HGNC:29676] HOMER homer scaffolding protein 2 [Source:HGNC Symbol;Acc:HGNC:17513] XRRA X-ray radiation resistance associated 1 [Source:HGNC Symbol;Acc:HGNC:18868] SNN 8303 stannin [Source:HGNC Symbol;Acc:HGNC:11149] MTERF mitochondrial transcription termination factor 2 [Source:HGNC Symbol;Acc:HGNC:30779] FGD FYVE, RhoGEF and PH domain containing 3 [Source:HGNC Symbol;Acc:HGNC:16027] CCDC coiled-coil domain containing 174 [Source:HGNC Symbol;Acc:HGNC:28033] GLT1D glycosyltransferase 1 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:26483] NKIRAS NFKB inhibitor interacting Ras like 1 [Source:HGNC Symbol;Acc:HGNC:17899] FERMT fermitin family member 3 [Source:HGNC Symbol;Acc:HGNC:23151] EGLN egl-9 family hypoxia inducible factor 2 [Source:HGNC Symbol;Acc:HGNC:14660] USP ubiquitin specific peptidase 48 [Source:HGNC Symbol;Acc:HGNC:18533] CNDP carnosine dipeptidase 1 [Source:HGNC Symbol;Acc:HGNC:20675] FAM65B 9750 family with sequence similarity 65 member B [Source:HGNC Symbol;Acc:HGNC:13872] LARP La ribonucleoprotein domain family member 1 [Source:HGNC Symbol;Acc:HGNC:29531] AQP aquaporin 10 [Source:HGNC Symbol;Acc:HGNC:16029] FAM168B family with sequence similarity 168 member B [Source:HGNC Symbol;Acc:HGNC:27016] TMEM200B transmembrane protein 200B [Source:HGNC Symbol;Acc:HGNC:33785] P2RY purinergic receptor P2Y12 [Source:HGNC Symbol;Acc:HGNC:18124] CLU 1191 clusterin [Source:HGNC Symbol;Acc:HGNC:2095] DDX3Y 8653 DEAD-box helicase 3, Y-linked [Source:HGNC Symbol;Acc:HGNC:2699] STXBP syntaxin binding protein 2 [Source:HGNC Symbol;Acc:HGNC:11445] AK4 205 adenylate kinase 4 [Source:HGNC Symbol;Acc:HGNC:363] BST2 684 bone marrow stromal cell antigen 2 [Source:HGNC Symbol;Acc:HGNC:1119] SRRM serine and arginine repetitive matrix 1 [Source:HGNC Symbol;Acc:HGNC:16638] KXD KxDL motif containing 1 [Source:HGNC Symbol;Acc:HGNC:28420] PABPC poly(a) binding protein cytoplasmic 3 [Source:HGNC Symbol;Acc:HGNC:8556] PSMD proteasome 26S subunit, non-atpase 4 [Source:HGNC Symbol;Acc:HGNC:9561] TBXA2R 6915 thromboxane A2 receptor [Source:HGNC Symbol;Acc:HGNC:11608] ZWINT ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] VAMP vesicle associated membrane protein 1 [Source:HGNC Symbol;Acc:HGNC:12642] HMHA Mitochondrial Calcium Uniporter RAD23A 5886 RAD23 homolog A, nucleotide excision repair protein [Source:HGNC Symbol;Acc:HGNC:9812] PIN peptidylprolyl cis/trans isomerase, NIMA-interacting 4 [Source:HGNC Symbol;Acc:HGNC:8992] DGAT diacylglycerol O-acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:2843] OVOS Ovostatin IQCJ IQ motif containing J [Source:HGNC Symbol;Acc:HGNC:32406] IQCJ-SCHIP IQCJ-SCHIP1 readthrough [Source:HGNC Symbol;Acc:HGNC:38842] SCHIP schwannomin interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:15678] PTPRJ 5795 protein tyrosine phosphatase, receptor type J [Source:HGNC Symbol;Acc:HGNC:9673] DGAT diacylglycerol O-acyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:16940] KCNN potassium calcium-activated channel subfamily N member 2 [Source:HGNC Symbol;Acc:HGNC:6291] CORO1A coronin 1A [Source:HGNC Symbol;Acc:HGNC:2252] UBE2N 7334 ubiquitin conjugating enzyme E2 N [Source:HGNC Symbol;Acc:HGNC:12492] TMEM transmembrane protein 97 [Source:HGNC Symbol;Acc:HGNC:28106] GAS2L growth arrest specific 2 like 1 [Source:HGNC Symbol;Acc:HGNC:16955] TNC 3371 tenascin C [Source:HGNC Symbol;Acc:HGNC:5318] SLC25A5 292 solute carrier family 25 member 5 [Source:HGNC Symbol;Acc:HGNC:10991] DNM1L dynamin 1-like [Source:HGNC Symbol;Acc:HGNC:2973] EIF1AY 9086 eukaryotic translation initiation factor 1A, Y-linked [Source:HGNC Symbol;Acc:HGNC:3252] HMOX heme oxygenase 1 [Source:HGNC Symbol;Acc:HGNC:5013] FKBP FK506 binding protein 3 [Source:HGNC Symbol;Acc:HGNC:3719] CD CD36 molecule [Source:HGNC Symbol;Acc:HGNC:1663] MAEA macrophage erythroblast attacher [Source:HGNC Symbol;Acc:HGNC:13731] DOK docking protein 3 [Source:HGNC Symbol;Acc:HGNC:24583] PXN 5829 paxillin [Source:HGNC Symbol;Acc:HGNC:9718] 4.2 2
13 374 NUDT nudix hydrolase 1 [Source:HGNC Symbol;Acc:HGNC:8048] PCGF polycomb group ring finger 3 [Source:HGNC Symbol;Acc:HGNC:10066] SDHB 6390 succinate dehydrogenase complex iron sulfur subunit B [Source:HGNC Symbol;Acc:HGNC:10681] SEMA4D semaphorin 4D [Source:HGNC Symbol;Acc:HGNC:10732] RNF ring finger protein 5 [Source:HGNC Symbol;Acc:HGNC:10068] HIST2H2BE 8349 histone cluster 2, H2be [Source:HGNC Symbol;Acc:HGNC:4760] IQGAP IQ motif containing GTPase activating protein 3 [Source:HGNC Symbol;Acc:HGNC:20669] PRPF pre-mrna processing factor 31 [Source:HGNC Symbol;Acc:HGNC:15446] FNTA 2339 farnesyltransferase, CAAX box, alpha [Source:HGNC Symbol;Acc:HGNC:3782] CCR C-C motif chemokine receptor 7 [Source:HGNC Symbol;Acc:HGNC:1608] RSL24D ribosomal L24 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:18479] UBE2L ubiquitin conjugating enzyme E2 L6 [Source:HGNC Symbol;Acc:HGNC:12490] ITGB integrin subunit beta 3 [Source:HGNC Symbol;Acc:HGNC:6156] PAGE PAGE family member 2 [Source:HGNC Symbol;Acc:HGNC:31804] PTCRA pre T-cell antigen receptor alpha [Source:HGNC Symbol;Acc:HGNC:21290] LCN lipocalin 2 [Source:HGNC Symbol;Acc:HGNC:6526] NBEAL neurobeachin like 2 [Source:HGNC Symbol;Acc:HGNC:31928] SUV420H Lysine Methyltransferase 5B ANKRD ankyrin repeat domain 28 [Source:HGNC Symbol;Acc:HGNC:29024] SCLY selenocysteine lyase [Source:HGNC Symbol;Acc:HGNC:18161] ALDH18A aldehyde dehydrogenase 18 family member A1 [Source:HGNC Symbol;Acc:HGNC:9722] SERPINE serpin family E member 1 [Source:HGNC Symbol;Acc:HGNC:8583] CEP centrosomal protein 57 [Source:HGNC Symbol;Acc:HGNC:30794] ZNF zinc finger protein 263 [Source:HGNC Symbol;Acc:HGNC:13056] TCTN tectonic family member 3 [Source:HGNC Symbol;Acc:HGNC:24519] SLC25A solute carrier family 25 member 10 [Source:HGNC Symbol;Acc:HGNC:10980] CRIM cysteine rich transmembrane BMP regulator 1 (chordin-like) [Source:HGNC Symbol;Acc:HGNC:2359] SLC11A solute carrier family 11 member 1 [Source:HGNC Symbol;Acc:HGNC:10907] FAM110A family with sequence similarity 110 member A [Source:HGNC Symbol;Acc:HGNC:16188] CCDC coiled-coil domain containing 58 [Source:HGNC Symbol;Acc:HGNC:31136] TMCO transmembrane and coiled-coil domains 1 [Source:HGNC Symbol;Acc:HGNC:18188] LGALSL galectin like [Source:HGNC Symbol;Acc:HGNC:25012] XRCC X-ray repair cross complementing 4 [Source:HGNC Symbol;Acc:HGNC:12831] TPM tropomyosin 1 (alpha) [Source:HGNC Symbol;Acc:HGNC:12010] CXCR C-X-C motif chemokine receptor 2 [Source:HGNC Symbol;Acc:HGNC:6027] MYL myosin light chain 9 [Source:HGNC Symbol;Acc:HGNC:15754] KRTCAP keratinocyte associated protein 3 [Source:HGNC Symbol;Acc:HGNC:28943] PHF PHD finger protein 6 [Source:HGNC Symbol;Acc:HGNC:18145] FPR formyl peptide receptor 1 [Source:HGNC Symbol;Acc:HGNC:3826] ZBTB zinc finger and BTB domain containing 44 [Source:HGNC Symbol;Acc:HGNC:25001] CD CD37 molecule [Source:HGNC Symbol;Acc:HGNC:1666] FAM162A family with sequence similarity 162 member A [Source:HGNC Symbol;Acc:HGNC:17865] MARCH membrane associated ring-ch-type finger 2 [Source:HGNC Symbol;Acc:HGNC:28038] STX syntaxin 11 [Source:HGNC Symbol;Acc:HGNC:11429] NOP NOP56 ribonucleoprotein [Source:HGNC Symbol;Acc:HGNC:15911] CLEC2B 9976 C-type lectin domain family 2 member B [Source:HGNC Symbol;Acc:HGNC:2053] RBCK RANBP2-type and C3HC4-type zinc finger containing 1 [Source:HGNC Symbol;Acc:HGNC:15864] CYB5A 1528 cytochrome b5 type A [Source:HGNC Symbol;Acc:HGNC:2570] METAP methionyl aminopeptidase 2 [Source:HGNC Symbol;Acc:HGNC:16672] ADCY7 113 adenylate cyclase 7 [Source:HGNC Symbol;Acc:HGNC:238] ESAM endothelial cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:17474] EPCAM 4072 epithelial cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:11529] BIRC5 332 baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] MEGF multiple EGF like domains 9 [Source:HGNC Symbol;Acc:HGNC:3234] DECR ,4-dienoyl-CoA reductase 1, mitochondrial [Source:HGNC Symbol;Acc:HGNC:2753] RNF ring finger protein 34 [Source:HGNC Symbol;Acc:HGNC:17297] PRPF40A pre-mrna processing factor 40 homolog A [Source:HGNC Symbol;Acc:HGNC:16463] CCL C-C motif chemokine ligand 5 [Source:HGNC Symbol;Acc:HGNC:10632] NFIC 4782 nuclear factor I C [Source:HGNC Symbol;Acc:HGNC:7786] SMIM small integral membrane protein 5 [Source:HGNC Symbol;Acc:HGNC:40030] PLAA 9373 phospholipase A2 activating protein [Source:HGNC Symbol;Acc:HGNC:9043] CR complement component 3b/4b receptor 1 (Knops blood group) [Source:HGNC Symbol;Acc:HGNC:2334] VNN vanin 2 [Source:HGNC Symbol;Acc:HGNC:12706] CENPH centromere protein H [Source:HGNC Symbol;Acc:HGNC:17268] VAPB 9217 VAMP (vesicle-associated membrane protein)-associated protein B and C [Source:HGNC Symbol;Acc:HGNC:12649] HBG hemoglobin subunit gamma 2 [Source:HGNC Symbol;Acc:HGNC:4832] DOK docking protein 2 [Source:HGNC Symbol;Acc:HGNC:2991] PCSK proprotein convertase subtilisin/kexin type 6 [Source:HGNC Symbol;Acc:HGNC:8569] HGD 3081 homogentisate 1,2-dioxygenase [Source:HGNC Symbol;Acc:HGNC:4892] GRINA 2907 glutamate ionotropic receptor NMDA type subunit associated protein 1 [Source:HGNC Symbol;Acc:HGNC:4589] PPP1R protein phosphatase 1 regulatory subunit 10 [Source:HGNC Symbol;Acc:HGNC:9284] ARHGAP6 395 Rho GTPase activating protein 6 [Source:HGNC Symbol;Acc:HGNC:676] ZDHHC zinc finger DHHC-type containing 9 [Source:HGNC Symbol;Acc:HGNC:18475] AKAP A-kinase anchoring protein 1 [Source:HGNC Symbol;Acc:HGNC:367] UNC119B unc-119 lipid binding chaperone B [Source:HGNC Symbol;Acc:HGNC:16488] VPS37C VPS37C, ESCRT-I subunit [Source:HGNC Symbol;Acc:HGNC:26097] 4.0 2
14 450 LYL lymphoblastic leukemia associated hematopoiesis regulator 1 [Source:HGNC Symbol;Acc:HGNC:6734] ERAP endoplasmic reticulum aminopeptidase 2 [Source:HGNC Symbol;Acc:HGNC:29499] IKZF IKAROS family zinc finger 1 [Source:HGNC Symbol;Acc:HGNC:13176] SELL 6402 selectin L [Source:HGNC Symbol;Acc:HGNC:10720] CBR carbonyl reductase 4 [Source:HGNC Symbol;Acc:HGNC:25891] CMTM CKLF like MARVEL transmembrane domain containing 2 [Source:HGNC Symbol;Acc:HGNC:19173] NAA N(alpha)-acetyltransferase 50, NatE catalytic subunit [Source:HGNC Symbol;Acc:HGNC:29533] LAMTOR late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 [Source:HGNC Symbol;Acc:HGNC:17955] CCDC coiled-coil domain containing 86 [Source:HGNC Symbol;Acc:HGNC:28359] HDAC histone deacetylase 3 [Source:HGNC Symbol;Acc:HGNC:4854] NPFFR neuropeptide FF receptor 2 [Source:HGNC Symbol;Acc:HGNC:4525] MLH mutl homolog 3 [Source:HGNC Symbol;Acc:HGNC:7128] CCDC coiled-coil domain containing 53 [Source:HGNC Symbol;Acc:HGNC:24256] GRB growth factor receptor bound protein 10 [Source:HGNC Symbol;Acc:HGNC:4564] EHD EH domain containing 3 [Source:HGNC Symbol;Acc:HGNC:3244] PREX phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 1 [Source:HGNC Symbol;Acc:HGNC:32594] GP glycoprotein VI platelet [Source:HGNC Symbol;Acc:HGNC:14388] HBS1L HBS1 like translational GTPase [Source:HGNC Symbol;Acc:HGNC:4834] HIPK homeodomain interacting protein kinase 2 [Source:HGNC Symbol;Acc:HGNC:14402] CBR1 873 carbonyl reductase 1 [Source:HGNC Symbol;Acc:HGNC:1548] BAZ1B 9031 bromodomain adjacent to zinc finger domain 1B [Source:HGNC Symbol;Acc:HGNC:961] SMIM small integral membrane protein 15 [Source:HGNC Symbol;Acc:HGNC:33861] UBE2K 3093 ubiquitin conjugating enzyme E2 K [Source:HGNC Symbol;Acc:HGNC:4914] PPFIA PTPRF interacting protein alpha 1 [Source:HGNC Symbol;Acc:HGNC:9245] MAP2K mitogen-activated protein kinase kinase 3 [Source:HGNC Symbol;Acc:HGNC:6843] PREPL 9581 prolyl endopeptidase-like [Source:HGNC Symbol;Acc:HGNC:30228] ARHGAP Rho GTPase activating protein 9 [Source:HGNC Symbol;Acc:HGNC:14130] OCIAD OCIA domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28685] BRD bromodomain containing 9 [Source:HGNC Symbol;Acc:HGNC:25818] NUDT nudix hydrolase 5 [Source:HGNC Symbol;Acc:HGNC:8052] DEPDC1B DEP domain containing 1B [Source:HGNC Symbol;Acc:HGNC:24902] TNFRSF10C 8794 tumor necrosis factor receptor superfamily member 10c [Source:HGNC Symbol;Acc:HGNC:11906] CT45A cancer/testis antigen family 45, member A1 [Source:HGNC Symbol;Acc:HGNC:33267] PIM Pim-3 proto-oncogene, serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:19310] SLC24A solute carrier family 24 member 4 [Source:HGNC Symbol;Acc:HGNC:10978] TCERG transcription elongation regulator 1 [Source:HGNC Symbol;Acc:HGNC:15630] HCAR hydroxycarboxylic acid receptor 3 [Source:HGNC Symbol;Acc:HGNC:16824] UBP upstream binding protein 1 (LBP-1a) [Source:HGNC Symbol;Acc:HGNC:12507] BET Bet1 golgi vesicular membrane trafficking protein [Source:HGNC Symbol;Acc:HGNC:14562] GAB GRB2 associated binding protein 1 [Source:HGNC Symbol;Acc:HGNC:4066] ATG16L autophagy related 16 like 2 [Source:HGNC Symbol;Acc:HGNC:25464] GSK3B 2932 glycogen synthase kinase 3 beta [Source:HGNC Symbol;Acc:HGNC:4617] IQSEC IQ motif and Sec7 domain 1 [Source:HGNC Symbol;Acc:HGNC:29112] CAV2 858 caveolin 2 [Source:HGNC Symbol;Acc:HGNC:1528] DNM dynamin 3 [Source:HGNC Symbol;Acc:HGNC:29125] BIN bridging integrator 2 [Source:HGNC Symbol;Acc:HGNC:1053] IL6R 3570 interleukin 6 receptor [Source:HGNC Symbol;Acc:HGNC:6019] VPS13A vacuolar protein sorting 13 homolog A [Source:HGNC Symbol;Acc:HGNC:1908] EIF5A 1984 eukaryotic translation initiation factor 5A [Source:HGNC Symbol;Acc:HGNC:3300] NUFIP NUFIP2, FMR1 interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:17634] PNRC proline rich nuclear receptor coactivator 2 [Source:HGNC Symbol;Acc:HGNC:23158] SMOX spermine oxidase [Source:HGNC Symbol;Acc:HGNC:15862] G6PD 2539 glucose-6-phosphate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4057] TWISTNB TWIST neighbor [Source:HGNC Symbol;Acc:HGNC:18027] MLXIP MLX interacting protein [Source:HGNC Symbol;Acc:HGNC:17055] DCAF DDB1 and CUL4 associated factor 5 [Source:HGNC Symbol;Acc:HGNC:20224] DDX19A DEAD-box helicase 19A [Source:HGNC Symbol;Acc:HGNC:25628] MYO1F 4542 myosin IF [Source:HGNC Symbol;Acc:HGNC:7600] MRPL mitochondrial ribosomal protein L43 [Source:HGNC Symbol;Acc:HGNC:14517] C6orf chromosome 6 open reading frame 25 [Source:HGNC Symbol;Acc:HGNC:13937] TMEM transmembrane protein 147 [Source:HGNC Symbol;Acc:HGNC:30414] BCAS breast carcinoma amplified sequence 3 [Source:HGNC Symbol;Acc:HGNC:14347] TRIB tribbles pseudokinase 1 [Source:HGNC Symbol;Acc:HGNC:16891] KIF2A 3796 kinesin heavy chain member 2A [Source:HGNC Symbol;Acc:HGNC:6318] TGFB transforming growth factor beta 1 [Source:HGNC Symbol;Acc:HGNC:11766] SUPT16H SPT16 homolog, facilitates chromatin remodeling subunit [Source:HGNC Symbol;Acc:HGNC:11465] ERP endoplasmic reticulum protein 44 [Source:HGNC Symbol;Acc:HGNC:18311] MYZAP myocardial zonula adherens protein [Source:HGNC Symbol;Acc:HGNC:43444] ARID1A 8289 AT-rich interaction domain 1A [Source:HGNC Symbol;Acc:HGNC:11110] TIMM translocase of inner mitochondrial membrane 10 homolog (yeast) [Source:HGNC Symbol;Acc:HGNC:11814] TRAPPC6B trafficking protein particle complex 6B [Source:HGNC Symbol;Acc:HGNC:23066] NOP NOP16 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:26934] CD CD96 molecule [Source:HGNC Symbol;Acc:HGNC:16892] HAL 3034 histidine ammonia-lyase [Source:HGNC Symbol;Acc:HGNC:4806] SSX SSX family member 2 [Source:HGNC Symbol;Acc:HGNC:11336] 3.8 2
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor
 Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
Table S2. Sequences immunoprecipitating with anti-hif-1α
 Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ ΡΑΣΗ ΤΟΥ ΓΑΣΤΡΙΚΟΥ ΒΛΕΝΝΟΓΟΝΟΥ ΣΤΗ ΛΟΙΜΩΞΗ ΜΕ ΕΛΙΚΟΒΑΚΤΗΡΙ ΙΟ ΤΟΥ ΠΥΛΩΡΟΥ ΠΡΙΝ ΚΑΙ ΜΕΤΑ ΤΗ ΘΕΡΑΠΕΙΑ
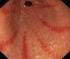 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo
 Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Supplemental Figures and Tables
 KLF2 and KLF4 control endothelial identity and vascular integrity Panjamaporn Sangwung 1, 2,#, Guangjin Zhou 1,#, Lalitha Nayak 1,3, E. Ricky Chan 4, Sandeep Kumar 5, Dong-Won Kang 5, Rongli Zhang 1, Xudong
KLF2 and KLF4 control endothelial identity and vascular integrity Panjamaporn Sangwung 1, 2,#, Guangjin Zhou 1,#, Lalitha Nayak 1,3, E. Ricky Chan 4, Sandeep Kumar 5, Dong-Won Kang 5, Rongli Zhang 1, Xudong
Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ ΩΝ ΚΑΤΑ ΤΗΝ ΕΜΒΡΥΪΚΗ ΑΝΑΠΤΥΞΗ
 ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Legends for Supplemental Data
 Legends for Supplemental Data Supplemental Fig. 1. Top molecular and cellular functions identified by Ingenuity Pathway Analysis after 6 h (left) and 24 h (right) of exposure of mouse pancreatic islets
Legends for Supplemental Data Supplemental Fig. 1. Top molecular and cellular functions identified by Ingenuity Pathway Analysis after 6 h (left) and 24 h (right) of exposure of mouse pancreatic islets
High mobility group 1 HMG1
 Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Hvordan påvirker CO 2 post-smolt i brakkvann RAS?
 Hvordan påvirker CO 2 post-smolt i brakkvann RAS? Tom O. Nilsen,. Mota, V.C., Kolarevic, J., Ytterborg, E., Ebbesson, Lars O.E., Handeland, S.O., Stiller, K., Terjesen, B.F SUNNDALSØRA, 12. NOVEMBER 2018
Hvordan påvirker CO 2 post-smolt i brakkvann RAS? Tom O. Nilsen,. Mota, V.C., Kolarevic, J., Ytterborg, E., Ebbesson, Lars O.E., Handeland, S.O., Stiller, K., Terjesen, B.F SUNNDALSØRA, 12. NOVEMBER 2018
Debashish Sahay. To cite this version: HAL Id: tel
 Identification of genes activated and biological markers involved in lysophosphatidic acid (LPA)-induced breast cancer metastasis through its receptor LPA1 Debashish Sahay To cite this version: Debashish
Identification of genes activated and biological markers involved in lysophosphatidic acid (LPA)-induced breast cancer metastasis through its receptor LPA1 Debashish Sahay To cite this version: Debashish
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated
 Supplemental Figure legends Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated genes. a) Mitotic roles of Polo-like kinase. b) Cell cycle regulation by BTG
Supplemental Figure legends Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated genes. a) Mitotic roles of Polo-like kinase. b) Cell cycle regulation by BTG
Supporting Information. Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka
 Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance
 POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
Table S1A. Generic EMT signature for tumour
 Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
ΙΩΑΝΝΗΣ ΚΛΑΓΚΑΣ. Χημικός, MSc ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ ΥΠΟΒΛΗΘΗΚΕ ΣΤΗΝ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΥ ΑΡΙΣΤΟΤΕΛΕΙΟΥ ΠΑΝΕΠΙΣΤΗΜΙΟΥ ΘΕΣΣΑΛΟΝΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΦΥΣΙΟΛΟΓΙΑΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ B ΕΡΓΑΣΤΗΡΙΟ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΔΙΕΥΘΥΝΤΗΣ: ΚΑΘΗΓΗΤΗΣ ΓΙΩΡΓΟΣ ΚΑΡΑΚΙΟΥΛΑΚΗΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 ΑΡΙΘΜ. 2376 Η ΕΠΙΔΡΑΣΗ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΦΥΣΙΟΛΟΓΙΑΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ B ΕΡΓΑΣΤΗΡΙΟ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΔΙΕΥΘΥΝΤΗΣ: ΚΑΘΗΓΗΤΗΣ ΓΙΩΡΓΟΣ ΚΑΡΑΚΙΟΥΛΑΚΗΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 ΑΡΙΘΜ. 2376 Η ΕΠΙΔΡΑΣΗ
Differentially expressed genes of HEK293/HeLa Myst4 sirna vs. controls.
 Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Figure S1. Localization of GiOR-1 with truncated N-terminal domain of 29 amino acid residues (ΔL-GiOR-1-HA).
 Figure S1. Localization of GiOR-1 with truncated N-terminal domain of 29 amino acid residues (ΔL-GiOR-1-HA). A L-GiOR-1-HA Tom40 DAPI merge DIC B LYS CYT ORG ORG Trypsin - - - + L-GiOR-1-HA Figure S2A.
Figure S1. Localization of GiOR-1 with truncated N-terminal domain of 29 amino acid residues (ΔL-GiOR-1-HA). A L-GiOR-1-HA Tom40 DAPI merge DIC B LYS CYT ORG ORG Trypsin - - - + L-GiOR-1-HA Figure S2A.
ΥΠΕΡΙΝΣΟΥΛΙΝΑΙΜΙΑ ΚΑΙ ΑΝΤΙΣΤΑΣΗ ΣΤΗΝ ΙΝΣΟΥΛΙΝΗ ΣΕ ΠΑΧΥΣΑΡΚΑ ΠΑΙΔΙΑ ΚΑΙ ΕΦΗΒΟΥΣ ΜΕ ΠΡΩΙΜΗ ΑΔΡΕΝΑΡΧΗ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΒΙΟΛΟΓΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΠΡΟΛΗΠΤΙΚΗΣ ΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΒΙΟΧΗΜΕΙΑΣ ΔΙΕΥΘΥΝΤΡΙΑ: Η ΚΑΘΗΓΗΤΡΙΑ Ν. ΒΑΒΑΤΣΗ-ΧΡΙΣΤΑΚΗ ΠΑΝΕΠ. ΕΤΟΣ 2009-2010 Αριθμ.2456
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΒΙΟΛΟΓΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΠΡΟΛΗΠΤΙΚΗΣ ΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΒΙΟΧΗΜΕΙΑΣ ΔΙΕΥΘΥΝΤΡΙΑ: Η ΚΑΘΗΓΗΤΡΙΑ Ν. ΒΑΒΑΤΣΗ-ΧΡΙΣΤΑΚΗ ΠΑΝΕΠ. ΕΤΟΣ 2009-2010 Αριθμ.2456
ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΣΕΞΟΥΑΛΙΚΗΣ ΔΡΑΣΤΗΡΙΟΤΗΤΑΣ ΤΩΝ ΓΥΝΑΙΚΩΝ ΚΑΤΑ ΤΗ ΔΙΑΡΚΕΙΑ ΤΗΣ ΕΓΚΥΜΟΣΥΝΗΣ ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΣΕΞΟΥΑΛΙΚΗΣ ΔΡΑΣΤΗΡΙΟΤΗΤΑΣ ΤΩΝ ΓΥΝΑΙΚΩΝ ΚΑΤΑ ΤΗ ΔΙΑΡΚΕΙΑ ΤΗΣ ΕΓΚΥΜΟΣΥΝΗΣ ΑΝΔΡΕΟΥ ΣΤΕΦΑΝΙΑ Λεμεσός 2012 i ii ΤΕΧΝΟΛΟΓΙΚΟ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΣΕΞΟΥΑΛΙΚΗΣ ΔΡΑΣΤΗΡΙΟΤΗΤΑΣ ΤΩΝ ΓΥΝΑΙΚΩΝ ΚΑΤΑ ΤΗ ΔΙΑΡΚΕΙΑ ΤΗΣ ΕΓΚΥΜΟΣΥΝΗΣ ΑΝΔΡΕΟΥ ΣΤΕΦΑΝΙΑ Λεμεσός 2012 i ii ΤΕΧΝΟΛΟΓΙΚΟ
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine
 Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL 1. Methods MicroRNA TaqMan Low Density Array MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Life Technologies). Global
SUPPLEMENTAL MATERIAL 1. Methods MicroRNA TaqMan Low Density Array MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Life Technologies). Global
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
RHOA inactivation enhances Wnt signaling and promotes colorectal cancer
 RHOA and colorectal cancer Paulo Rodrigues et al 2014 SUPPLEMENTARY INFORMATION RHOA inactivation enhances Wnt signaling and promotes colorectal cancer Paulo Rodrigues, Irati Macaya, Sarah Bazzocco, Rocco
RHOA and colorectal cancer Paulo Rodrigues et al 2014 SUPPLEMENTARY INFORMATION RHOA inactivation enhances Wnt signaling and promotes colorectal cancer Paulo Rodrigues, Irati Macaya, Sarah Bazzocco, Rocco
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Linking FOXO3, NCOA3, and TCF7L2 to Ras pathway phenotypes through a genomewide forward genetic screen in human colorectal cancer cells
 1 2 3 4 5 6 7 8 9 Linking FOXO3, NCOA3, and TCF7L2 to Ras pathway phenotypes through a genomewide forward genetic screen in human colorectal cancer cells Authors: Snehangshu Kundu 1, Muhammad Akhtar Ali
1 2 3 4 5 6 7 8 9 Linking FOXO3, NCOA3, and TCF7L2 to Ras pathway phenotypes through a genomewide forward genetic screen in human colorectal cancer cells Authors: Snehangshu Kundu 1, Muhammad Akhtar Ali
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
A rearrangement is established as a barrier in parapatry. The accumulation of incompatibilites starts. Chrom 1. Rearranged
 Figure S1. An ancestral species splits in two according to the model discussed in Navarro and Barton (Evolution. In press. 2003). A rearrangement in chromosome 1 was involved in the process. From the moment
Figure S1. An ancestral species splits in two according to the model discussed in Navarro and Barton (Evolution. In press. 2003). A rearrangement in chromosome 1 was involved in the process. From the moment
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Κυτταρική Επικοινωνία Είναι η ικανότητα του κυττάρου να αποκρίνεται σε εξωτερικά σήματα (σήματα από το περιβάλλον του ή άλλα κύτταρα) Τα σήματα αυτά μπορεί να είναι
Κυτταρική Επικοινωνία Cell Communication Κυτταρική Επικοινωνία Είναι η ικανότητα του κυττάρου να αποκρίνεται σε εξωτερικά σήματα (σήματα από το περιβάλλον του ή άλλα κύτταρα) Τα σήματα αυτά μπορεί να είναι
Gene Primer sequence (5 to 3 ) Size (bp) E (%) GenBank
 Table S1 Primers used for quantitative real time PCR Gene Primer sequence (5 to 3 ) Size (bp) E (%) GenBank accession no: scd a got2a a ube2h a gpx4b a mknk2b a hsd11b2 b eef1a c b2m d F: ACCCGGAAGTCATCGAGAGA
Table S1 Primers used for quantitative real time PCR Gene Primer sequence (5 to 3 ) Size (bp) E (%) GenBank accession no: scd a got2a a ube2h a gpx4b a mknk2b a hsd11b2 b eef1a c b2m d F: ACCCGGAAGTCATCGAGAGA
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΒΙΟΧΗΜΕΙΑΣ & ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test Επιμέλεια
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΒΙΟΧΗΜΕΙΑΣ & ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test Επιμέλεια
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
1) Abstract (To be organized as: background, aim, workpackages, expected results) (300 words max) Το όριο λέξεων θα είναι ελαστικό.
 UΓενικές Επισημάνσεις 1. Παρακάτω θα βρείτε απαντήσεις του Υπουργείου, σχετικά με τη συμπλήρωση της ηλεκτρονικής φόρμας. Διευκρινίζεται ότι στα περισσότερα θέματα οι απαντήσεις ήταν προφορικές (τηλεφωνικά),
UΓενικές Επισημάνσεις 1. Παρακάτω θα βρείτε απαντήσεις του Υπουργείου, σχετικά με τη συμπλήρωση της ηλεκτρονικής φόρμας. Διευκρινίζεται ότι στα περισσότερα θέματα οι απαντήσεις ήταν προφορικές (τηλεφωνικά),
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
Σχέση στεφανιαίας νόσου και άγχους - κατάθλιψης
 Τρίμηνη, ηλεκτρονική έκδοση του Τμήματος Νοσηλευτικής Α, Τεχνολογικό Εκπαιδευτικό Ίδρυμα Αθήνας _ΑΝΑΣΚΟΠΗΣΗ_ Πολυκανδριώτη Μαρία 1, Φούκα Γεωργία 2 1. Καθηγήτρια Εφαρμογών Νοσηλευτικής Α, ΤΕΙ Αθήνας 2.
Τρίμηνη, ηλεκτρονική έκδοση του Τμήματος Νοσηλευτικής Α, Τεχνολογικό Εκπαιδευτικό Ίδρυμα Αθήνας _ΑΝΑΣΚΟΠΗΣΗ_ Πολυκανδριώτη Μαρία 1, Φούκα Γεωργία 2 1. Καθηγήτρια Εφαρμογών Νοσηλευτικής Α, ΤΕΙ Αθήνας 2.
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία "Η ΣΗΜΑΝΤΙΚΟΤΗΤΑ ΤΟΥ ΜΗΤΡΙΚΟΥ ΘΗΛΑΣΜΟΥ ΣΤΗ ΠΡΟΛΗΨΗ ΤΗΣ ΠΑΙΔΙΚΗΣ ΠΑΧΥΣΑΡΚΙΑΣ" Ειρήνη Σωτηρίου Λεμεσός 2014 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία "Η ΣΗΜΑΝΤΙΚΟΤΗΤΑ ΤΟΥ ΜΗΤΡΙΚΟΥ ΘΗΛΑΣΜΟΥ ΣΤΗ ΠΡΟΛΗΨΗ ΤΗΣ ΠΑΙΔΙΚΗΣ ΠΑΧΥΣΑΡΚΙΑΣ" Ειρήνη Σωτηρίου Λεμεσός 2014 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
Supporting Information
 Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were
Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were
ΜΗΤΡΙΚΟΣ ΘΗΛΑΣΜΟΣ ΚΑΙ ΓΝΩΣΤΙΚΗ ΑΝΑΠΤΥΞΗ ΜΕΧΡΙ ΚΑΙ 10 ΧΡΟΝΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΜΗΤΡΙΚΟΣ ΘΗΛΑΣΜΟΣ ΚΑΙ ΓΝΩΣΤΙΚΗ ΑΝΑΠΤΥΞΗ ΜΕΧΡΙ ΚΑΙ 10 ΧΡΟΝΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Ονοματεπώνυμο Κεντούλλα Πέτρου Αριθμός Φοιτητικής Ταυτότητας 2008761539 Κύπρος
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΜΗΤΡΙΚΟΣ ΘΗΛΑΣΜΟΣ ΚΑΙ ΓΝΩΣΤΙΚΗ ΑΝΑΠΤΥΞΗ ΜΕΧΡΙ ΚΑΙ 10 ΧΡΟΝΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Ονοματεπώνυμο Κεντούλλα Πέτρου Αριθμός Φοιτητικής Ταυτότητας 2008761539 Κύπρος
New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp.
 SUPPLEMENTARY MATERIAL New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp. Mohamed-Elamir F. Hegazy a,*, Amira M. Gamal-Eldeen b, Tarik A. Mohamed a, Montaser A. Alhammady c, Abuzeid A.
SUPPLEMENTARY MATERIAL New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp. Mohamed-Elamir F. Hegazy a,*, Amira M. Gamal-Eldeen b, Tarik A. Mohamed a, Montaser A. Alhammady c, Abuzeid A.
Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver
 Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Abstract... I. Zusammenfassung... II. 1 Aim of the work Introduction Short overview of Chinese hamster ovary cell lines...
 III Contents I II III Abstract... I Zusammenfassung... II Contents... III 1 Aim of the work... 1 2 Introduction... 2 2.1 Short overview of Chinese hamster ovary cell lines... 2 2.2 Unspecific mutations:
III Contents I II III Abstract... I Zusammenfassung... II Contents... III 1 Aim of the work... 1 2 Introduction... 2 2.1 Short overview of Chinese hamster ovary cell lines... 2 2.2 Unspecific mutations:
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Splice site recognition between different organisms
 NATIONAL AND KAPODISTRIAN UNIVERSITY OF ATHENS SCHOOL OF SCIENCE DEPARTMENT OF INFORMATICS AND TELECOMMUNICATIONS INTERDEPARTMENTAL POSTGRADUATE PROGRAM "INFORMATION TECHNOLOGIES IN MEDICINE AND BIOLOGY"
NATIONAL AND KAPODISTRIAN UNIVERSITY OF ATHENS SCHOOL OF SCIENCE DEPARTMENT OF INFORMATICS AND TELECOMMUNICATIONS INTERDEPARTMENTAL POSTGRADUATE PROGRAM "INFORMATION TECHNOLOGIES IN MEDICINE AND BIOLOGY"
 Supplemental Figure 1 Genechip data analysis. Three pairs of bulge and sub-bulge ORS Genechips were analyzed and compared by MASv5.0 and dchip 1.3 software algorithms in order to identify gene transcripts
Supplemental Figure 1 Genechip data analysis. Three pairs of bulge and sub-bulge ORS Genechips were analyzed and compared by MASv5.0 and dchip 1.3 software algorithms in order to identify gene transcripts
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ Παθοφυσιολογία ΙΙ Ογκολογία Υπεύθυνος μαθήματος: Καθηγητής Αλέξανδρος Α. Δρόσος Άδειες Χρήσης Το παρόν εκπαιδευτικό υλικό υπόκειται σε άδειες χρήσης Creative
ΠΑΝΕΠΙΣΤΗΜΙΟ ΙΩΑΝΝΙΝΩΝ ΑΝΟΙΚΤΑ ΑΚΑΔΗΜΑΪΚΑ ΜΑΘΗΜΑΤΑ Παθοφυσιολογία ΙΙ Ογκολογία Υπεύθυνος μαθήματος: Καθηγητής Αλέξανδρος Α. Δρόσος Άδειες Χρήσης Το παρόν εκπαιδευτικό υλικό υπόκειται σε άδειες χρήσης Creative
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ Πτυχιακή εργασία ΓΝΩΣΕΙΣ ΚΑΙ ΣΤΑΣΕΙΣ ΝΟΣΗΛΕΥΤΩΝ ΠΡΟΣ ΤΟΥΣ ΦΟΡΕΙΣ ΜΕ ΣΥΝΔΡΟΜΟ ΕΠΙΚΤΗΤΗΣ ΑΝΟΣΟΑΝΕΠΑΡΚΕΙΑΣ (AIDS) Αλέξης Δημήτρη Α.Φ.Τ: 20085675385 Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ Πτυχιακή εργασία ΓΝΩΣΕΙΣ ΚΑΙ ΣΤΑΣΕΙΣ ΝΟΣΗΛΕΥΤΩΝ ΠΡΟΣ ΤΟΥΣ ΦΟΡΕΙΣ ΜΕ ΣΥΝΔΡΟΜΟ ΕΠΙΚΤΗΤΗΣ ΑΝΟΣΟΑΝΕΠΑΡΚΕΙΑΣ (AIDS) Αλέξης Δημήτρη Α.Φ.Τ: 20085675385 Λεμεσός
Introduction to Bioinformatics
 Introduction to Bioinformatics 260.602.01 September 2, 2005 Jonathan Pevsner, Ph.D. pevsner@jhmi.edu bioinformatics medical informatics Tool-users public health informatics databases algorithms Tool-makers
Introduction to Bioinformatics 260.602.01 September 2, 2005 Jonathan Pevsner, Ph.D. pevsner@jhmi.edu bioinformatics medical informatics Tool-users public health informatics databases algorithms Tool-makers
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΕΠΗΡΕΑΖΕΙ ΤΗΝ ΠΡΟΛΗΨΗ ΚΑΡΚΙΝΟΥ ΤΟΥ ΜΑΣΤΟΥ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΠΩΣ Η ΚΑΤΑΝΑΛΩΣΗ ΦΡΟΥΤΩΝ ΚΑΙ ΛΑΧΑΝΙΚΩΝ ΕΠΗΡΕΑΖΕΙ ΤΗΝ ΠΡΟΛΗΨΗ ΚΑΡΚΙΝΟΥ ΤΟΥ ΜΑΣΤΟΥ Όνομα φοιτήτριας ΚΑΛΑΠΟΔΑ ΜΑΡΚΕΛΛΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΠΩΣ Η ΚΑΤΑΝΑΛΩΣΗ ΦΡΟΥΤΩΝ ΚΑΙ ΛΑΧΑΝΙΚΩΝ ΕΠΗΡΕΑΖΕΙ ΤΗΝ ΠΡΟΛΗΨΗ ΚΑΡΚΙΝΟΥ ΤΟΥ ΜΑΣΤΟΥ Όνομα φοιτήτριας ΚΑΛΑΠΟΔΑ ΜΑΡΚΕΛΛΑ
ΑΝΩΤΑΤΟ ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΙΟΝΙΩΝ ΝΗΣΩΝ ΠΑΡΑΡΤΗΜΑ ΛΕΥΚΑΔΑΣ ΤΜΗΜΑ ΕΦΑΡΜΟΓΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ ΣΤΗ ΔΙΟΙΚΗΣΗ ΚΑΙ ΣΤΗΝ ΟΙΚΟΝΟΜΙΑ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ
 ΑΝΩΤΑΤΟ ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΙΟΝΙΩΝ ΝΗΣΩΝ ΠΑΡΑΡΤΗΜΑ ΛΕΥΚΑΔΑΣ ΤΜΗΜΑ ΕΦΑΡΜΟΓΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ ΣΤΗ ΔΙΟΙΚΗΣΗ ΚΑΙ ΣΤΗΝ ΟΙΚΟΝΟΜΙΑ Συστήματα ηλεκτρονικών κρατήσεων σε ξενοδοχειακές επιχειρήσεις και εφαρμογή
ΑΝΩΤΑΤΟ ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΙΟΝΙΩΝ ΝΗΣΩΝ ΠΑΡΑΡΤΗΜΑ ΛΕΥΚΑΔΑΣ ΤΜΗΜΑ ΕΦΑΡΜΟΓΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ ΣΤΗ ΔΙΟΙΚΗΣΗ ΚΑΙ ΣΤΗΝ ΟΙΚΟΝΟΜΙΑ Συστήματα ηλεκτρονικών κρατήσεων σε ξενοδοχειακές επιχειρήσεις και εφαρμογή
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
Ex4-MS2. Exon 4. pcp Pre-mRNA Pre-mRNA
 A -MS2 A Exon 4 B N.E. N.E. pcp + + - + + - C N.E. N.E. pcp + + - + + - Pre-mRNA Pre-mRNA U2 U1 U4 U5 U6 U2 U1 U4 U5 U6 1 2 3 4 5 6 1 2 3 4 5 6 Supplementary Figure 1. The and complexes contain U1 and
A -MS2 A Exon 4 B N.E. N.E. pcp + + - + + - C N.E. N.E. pcp + + - + + - Pre-mRNA Pre-mRNA U2 U1 U4 U5 U6 U2 U1 U4 U5 U6 1 2 3 4 5 6 1 2 3 4 5 6 Supplementary Figure 1. The and complexes contain U1 and
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
NATIONAL AND KAPODISTRIAN UNIVERSITY OF ATHENS SCHOOL OF SCIENCE FACULTY OF INFORMATICS AND TELECOMMUNICATIONS
 NATIONAL AND KAPODISTRIAN UNIVERSITY OF ATHENS SCHOOL OF SCIENCE FACULTY OF INFORMATICS AND TELECOMMUNICATIONS INTERDICIPLINARY POSTGRADUATE PROGRAMME "INFORMATION TECHNOLOGIES IN MEDICINE AND BIOLOGY"
NATIONAL AND KAPODISTRIAN UNIVERSITY OF ATHENS SCHOOL OF SCIENCE FACULTY OF INFORMATICS AND TELECOMMUNICATIONS INTERDICIPLINARY POSTGRADUATE PROGRAMME "INFORMATION TECHNOLOGIES IN MEDICINE AND BIOLOGY"
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
19 Ενδοκρινολόγος. Φάρµακα που στοχεύουν το β- κύτταρο ΕΥΡΥΔΙΚΗ ΠΑΠΑΔΟΠΟΥΛΟΥ ΕΙΣΑΓΩΓΗ ΜΗΧΑΝΙΣΜΟΣ ΔΡΑΣΗΣ
 Φάρµακα που στοχεύουν το β- κύτταρο ΕΥΡΥΔΙΚΗ ΠΑΠΑΔΟΠΟΥΛΟΥ 19 Ενδοκρινολόγος ΕΙΣΑΓΩΓΗ Οι σουλφονυλουρίες είναι η πρώτη κατηγορία υπογλυκαιμικών φαρμάκων που χορηγήθηκαν για την αντιμετώπιση του διαβήτη
Φάρµακα που στοχεύουν το β- κύτταρο ΕΥΡΥΔΙΚΗ ΠΑΠΑΔΟΠΟΥΛΟΥ 19 Ενδοκρινολόγος ΕΙΣΑΓΩΓΗ Οι σουλφονυλουρίες είναι η πρώτη κατηγορία υπογλυκαιμικών φαρμάκων που χορηγήθηκαν για την αντιμετώπιση του διαβήτη
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία Η ΣΧΕΣΗ ΤΗΣ ΠΑΧΥΣΑΡΚΙΑΣ ΜΕ ΤΟ ΣΑΚΧΑΡΩΔΗ ΔΙΒΗΤΗ ΚΥΗΣΗΣ Χρυστάλλα, Γεωργίου Λεμεσός 2014 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία Η ΣΧΕΣΗ ΤΗΣ ΠΑΧΥΣΑΡΚΙΑΣ ΜΕ ΤΟ ΣΑΚΧΑΡΩΔΗ ΔΙΒΗΤΗ ΚΥΗΣΗΣ Χρυστάλλα, Γεωργίου Λεμεσός 2014 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
athanasiadis@rhodes.aegean.gr , -.
 παιδαγωγικά ρεύµατα στο Αιγαίο Προσκήνιο 88 - * athanasiadis@rhodes.aegean.gr -., -.. Abstract The aim of this survey is to show how students of the three last school classes of the Primary School evaluated
παιδαγωγικά ρεύµατα στο Αιγαίο Προσκήνιο 88 - * athanasiadis@rhodes.aegean.gr -., -.. Abstract The aim of this survey is to show how students of the three last school classes of the Primary School evaluated
Μηχανισμοί πρόβλεψης προσήμων σε προσημασμένα μοντέλα κοινωνικών δικτύων ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΕΠΙΚΟΙΝΩΝΙΩΝ, ΗΛΕΚΤΡΟΝΙΚΗΣ ΚΑΙ ΣΥΣΤΗΜΑΤΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ Μηχανισμοί πρόβλεψης προσήμων σε προσημασμένα μοντέλα κοινωνικών
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΕΠΙΚΟΙΝΩΝΙΩΝ, ΗΛΕΚΤΡΟΝΙΚΗΣ ΚΑΙ ΣΥΣΤΗΜΑΤΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ Μηχανισμοί πρόβλεψης προσήμων σε προσημασμένα μοντέλα κοινωνικών
AΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ
 AΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΕΙΔΙΚΕΥΣΗΣ ΠΡΟΣΤΑΣΙΑ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΚΑΙ ΒΙΩΣΙΜΗ ΑΝΑΠΤΥΞΗ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΙΕΣΕΩΝ ΣΤΟ ΠΕΡΙΒΑΛΛΟΝ
AΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΕΙΔΙΚΕΥΣΗΣ ΠΡΟΣΤΑΣΙΑ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΚΑΙ ΒΙΩΣΙΜΗ ΑΝΑΠΤΥΞΗ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΙΕΣΕΩΝ ΣΤΟ ΠΕΡΙΒΑΛΛΟΝ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ. Πτυχιακή εργασία Η ΚΑΤΑΘΛΙΨΗ ΣΕ ΕΦΗΒΟΥΣ ΜΕ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΤΥΠΟΥ 1
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή εργασία Η ΚΑΤΑΘΛΙΨΗ ΣΕ ΕΦΗΒΟΥΣ ΜΕ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΤΥΠΟΥ 1 ΑΝΔΡΕΑΣ ΑΝΔΡΕΟΥ Φ.Τ:2008670839 Λεμεσός 2014 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή εργασία Η ΚΑΤΑΘΛΙΨΗ ΣΕ ΕΦΗΒΟΥΣ ΜΕ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΤΥΠΟΥ 1 ΑΝΔΡΕΑΣ ΑΝΔΡΕΟΥ Φ.Τ:2008670839 Λεμεσός 2014 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ
Supporting Information
 Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
(1) Describe the process by which mercury atoms become excited in a fluorescent tube (3)
 Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Figures Supplementary Figure 1. Tandem MS of yeast eif2b.
 Figures Supplementary Figure 1. Tandem MS of yeast eif2b. Tandem MS of the 56+ charge state (shaded blue) of the eif2b dimer showing that two α subunits ( at 2000 m/z) dissociate from the dimer when collision
Figures Supplementary Figure 1. Tandem MS of yeast eif2b. Tandem MS of the 56+ charge state (shaded blue) of the eif2b dimer showing that two α subunits ( at 2000 m/z) dissociate from the dimer when collision
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
Structural and Biochemical Studies of Cotranslational Protein Transport across the Endoplasmic Reticulum
 Structural and Biochemical Studies of Cotranslational Protein Transport across the Endoplasmic Reticulum Inaugural-Dissertation zur Erlangung des akademischen Grades Doktor der Naturwissenschaften - Dr.
Structural and Biochemical Studies of Cotranslational Protein Transport across the Endoplasmic Reticulum Inaugural-Dissertation zur Erlangung des akademischen Grades Doktor der Naturwissenschaften - Dr.
ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ
 ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΚΑΛΛΙΟΠΗ ΚΩΤΣΑ ΕΠΙΚ. ΚΑΘΗΓΗΤΡΙΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ Α.Π.Θ ΤΜΗΜΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ-ΔΙΑΒΗΤΟΛΟΓΙΚΟ
ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΚΑΛΛΙΟΠΗ ΚΩΤΣΑ ΕΠΙΚ. ΚΑΘΗΓΗΤΡΙΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ Α.Π.Θ ΤΜΗΜΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ-ΔΙΑΒΗΤΟΛΟΓΙΚΟ
Développement de virus HSV-1 (virus de l herpes simplex de type 1) oncolytiques ciblés pour traiter les carcinomes hépatocellulaires
 Développement de virus HSV-1 (virus de l herpes simplex de type 1) oncolytiques ciblés pour traiter les carcinomes hépatocellulaires Aldo Decio Pourchet To cite this version: Aldo Decio Pourchet. Développement
Développement de virus HSV-1 (virus de l herpes simplex de type 1) oncolytiques ciblés pour traiter les carcinomes hépatocellulaires Aldo Decio Pourchet To cite this version: Aldo Decio Pourchet. Développement
Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil
 J. Jpn. Soc. Soil Phys. No. +*0, p.- +*,**1 Eh * ** Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil Daisuke MURAKAMI* and Tatsuaki KASUBUCHI** * The United Graduate
J. Jpn. Soc. Soil Phys. No. +*0, p.- +*,**1 Eh * ** Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil Daisuke MURAKAMI* and Tatsuaki KASUBUCHI** * The United Graduate
Παιδί βαρήκοο ή με διαταραχή στο φάσμα του αυτισμού;
 ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ / ORIGINAL ARTICLE Παιδί βαρήκοο ή με διαταραχή στο φάσμα του αυτισμού; Hard of hearing child or suffering from autism spectrum disorder? Χειμώνα Θ 1 Βλατάκη Μ 2 Πρώιμος Ε 1 Παπουτσάκη
ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ / ORIGINAL ARTICLE Παιδί βαρήκοο ή με διαταραχή στο φάσμα του αυτισμού; Hard of hearing child or suffering from autism spectrum disorder? Χειμώνα Θ 1 Βλατάκη Μ 2 Πρώιμος Ε 1 Παπουτσάκη
Supporting information. An unusual bifunctional Tb-MOF for highly sensing of Ba 2+ ions and remarkable selectivities of CO 2 /N 2 and CO 2 /CH 4
 Electronic Supplementary Material (ESI) for Journal of Materials Chemistry A. This journal is The Royal Society of Chemistry 2015 Supporting information An unusual bifunctional Tb-MOF for highly sensing
Electronic Supplementary Material (ESI) for Journal of Materials Chemistry A. This journal is The Royal Society of Chemistry 2015 Supporting information An unusual bifunctional Tb-MOF for highly sensing
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
Engineering Tunable Single and Dual Optical. Emission from Ru(II)-Polypyridyl Complexes. Through Excited State Design
 Engineering Tunable Single and Dual Optical Emission from Ru(II)-Polypyridyl Complexes Through Excited State Design Supplementary Information Julia Romanova 1, Yousif Sadik 1, M. R. Ranga Prabhath 1,,
Engineering Tunable Single and Dual Optical Emission from Ru(II)-Polypyridyl Complexes Through Excited State Design Supplementary Information Julia Romanova 1, Yousif Sadik 1, M. R. Ranga Prabhath 1,,
The NF-κB pathways in connective tissue diseases
 κ The NF-κB pathways in connective tissue diseases TETSUO KUBOTA (Associate Professor) Tokyo Medical and Dental University Graduate School of Health Care Sciences NF-κB nuclear factor-kappa B NF-κB in
κ The NF-κB pathways in connective tissue diseases TETSUO KUBOTA (Associate Professor) Tokyo Medical and Dental University Graduate School of Health Care Sciences NF-κB nuclear factor-kappa B NF-κB in
Supporting information. Influence of Aerosol Acidity on the Chemical Composition of Secondary Organic Aerosol from β caryophyllene
 Supporting information Influence of Aerosol Acidity on the Chemical Composition of Secondary Organic Aerosol from β caryophyllene M. N. Chan 1, J. D. Surratt 2,*, A. W. H. Chan 2,**, K. Schilling 2, J.
Supporting information Influence of Aerosol Acidity on the Chemical Composition of Secondary Organic Aerosol from β caryophyllene M. N. Chan 1, J. D. Surratt 2,*, A. W. H. Chan 2,**, K. Schilling 2, J.
Μελέτη της επίδρασης αντικαρκινικών χημειοθεραπευτικών φαρμάκων στη ρύθμιση του Cdt1
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραμμα Μεταπτυχιακών Σπουδών Εφαρμογές Στις Βασικές Επιστήμες Κατεύθυνση: Φαρμακοκινητική Τοξικολογία Διπλωματική Εργασία Μελέτη της επίδρασης
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραμμα Μεταπτυχιακών Σπουδών Εφαρμογές Στις Βασικές Επιστήμες Κατεύθυνση: Φαρμακοκινητική Τοξικολογία Διπλωματική Εργασία Μελέτη της επίδρασης
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supporting Information
 Supporting Information Molecular Cage Impregnated Palladium Nanoparticles: Efficient, Additive-Free Heterogeneous Catalysts for Cyanation of Aryl Halides. Bijnaneswar Mondal, Koushik Acharyya, Prodip Howlader
Supporting Information Molecular Cage Impregnated Palladium Nanoparticles: Efficient, Additive-Free Heterogeneous Catalysts for Cyanation of Aryl Halides. Bijnaneswar Mondal, Koushik Acharyya, Prodip Howlader
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ. Πτυχιακή Εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία Η ΕΠΑΓΓΕΛΜΑΤΙΚΗ ΕΞΟΥΘΕΝΩΣΗ ΠΟΥ ΒΙΩΝΕΙ ΤΟ ΝΟΣΗΛΕΥΤΙΚΟ ΠΡΟΣΩΠΙΚΟ ΣΤΙΣ ΜΟΝΑΔΕΣ ΕΝΑΤΙΚΗΣ ΘΕΡΑΠΕΙΑΣ Άντρη Αγαθαγγέλου Λεμεσός 2012 i ΤΕΧΝΟΛΟΓΙΚΟ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Πτυχιακή Εργασία Η ΕΠΑΓΓΕΛΜΑΤΙΚΗ ΕΞΟΥΘΕΝΩΣΗ ΠΟΥ ΒΙΩΝΕΙ ΤΟ ΝΟΣΗΛΕΥΤΙΚΟ ΠΡΟΣΩΠΙΚΟ ΣΤΙΣ ΜΟΝΑΔΕΣ ΕΝΑΤΙΚΗΣ ΘΕΡΑΠΕΙΑΣ Άντρη Αγαθαγγέλου Λεμεσός 2012 i ΤΕΧΝΟΛΟΓΙΚΟ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ
 5 ο ΠΑΝΕΛΛΗΝΙΟ ΣΥΝΕΔΡΙΟ ΕΛΛΗΝΙΚΗΣ ΕΤΑΙΡΕΙΑΣ ΑΙΜΑΦΑΙΡΕΣΗΣ ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ Θωμάς Τσαγανός, MD, PhD Πανεπιστημιακός Υπότροφος Δ Παθολογική Κλινική Πανεπιστημιακό Γενικό Νοσοκομείο
5 ο ΠΑΝΕΛΛΗΝΙΟ ΣΥΝΕΔΡΙΟ ΕΛΛΗΝΙΚΗΣ ΕΤΑΙΡΕΙΑΣ ΑΙΜΑΦΑΙΡΕΣΗΣ ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ Θωμάς Τσαγανός, MD, PhD Πανεπιστημιακός Υπότροφος Δ Παθολογική Κλινική Πανεπιστημιακό Γενικό Νοσοκομείο
rp, ribosomal protein 60S rp mrna rpl30 rps14 RT-PCR mrna nonsense-mediated mrna decay smg-2 smg-2 mrna rp rpl-1 rpl-1
 rp, ribosomal protein 60S 47 rpl 40S 32 rps rp mrna rpl30 rps14 rpl12 rpl3 rps13 rp mrna nonsense-mediated mrna decay (NMD) C. elegans smg-2 smg-2 mrna 50 rp 7 rp 4 rpl 4 rp NMD mrna 6 rp mrna rp mrna
rp, ribosomal protein 60S 47 rpl 40S 32 rps rp mrna rpl30 rps14 rpl12 rpl3 rps13 rp mrna nonsense-mediated mrna decay (NMD) C. elegans smg-2 smg-2 mrna 50 rp 7 rp 4 rpl 4 rp NMD mrna 6 rp mrna rp mrna
Δυσκολίες που συναντούν οι μαθητές της Στ Δημοτικού στην κατανόηση της λειτουργίας του Συγκεντρωτικού Φακού
 ΜΟΥΡΑΤΙΔΗΣ ΧΑΡΑΛΑΜΠΟΣ Δυσκολίες που συναντούν οι μαθητές της Στ Δημοτικού στην κατανόηση της λειτουργίας του Συγκεντρωτικού Φακού Μεταπτυχιακή Εργασία Ειδίκευσης που υποβλήθηκε στο πλαίσιο του Προγράμματος
ΜΟΥΡΑΤΙΔΗΣ ΧΑΡΑΛΑΜΠΟΣ Δυσκολίες που συναντούν οι μαθητές της Στ Δημοτικού στην κατανόηση της λειτουργίας του Συγκεντρωτικού Φακού Μεταπτυχιακή Εργασία Ειδίκευσης που υποβλήθηκε στο πλαίσιο του Προγράμματος
