Supplemental Figures and Tables
|
|
|
- Σέργιος Παπάζογλου
- 6 χρόνια πριν
- Προβολές:
Transcript
1 KLF2 and KLF4 control endothelial identity and vascular integrity Panjamaporn Sangwung 1, 2,#, Guangjin Zhou 1,#, Lalitha Nayak 1,3, E. Ricky Chan 4, Sandeep Kumar 5, Dong-Won Kang 5, Rongli Zhang 1, Xudong Liao 1, Yuan Lu 1, Keiki Sugi 1, Hisashi Fujioka 7, Hong Shi 1, Stephanie D Lapping 1, Chandra C. Ghosh 8, Sarah J. Higgins 8, Samir M. Parikh 8, Hanjoon Jo 5,6, and Mukesh K Jain 1,2,9 Supplemental Figures and Tables 1
2 Supplemental Figure 1 Day 6 post-tamoxifen Day 5 post-tamoxifen Day 4 post-tamoxifen A Relative mrna expression B Relative mrna expression C Relative mrna expression D CRE Klf2 ** NS ** CRE EC-Klf2-KO EC-Klf4-KO EC-DKO KLF4 Klf4 NS ** ** Nos3 Thbd ** NS NS Klf2 Klf4 Nos3 Thbd ** 3 ** 3 ** 3 ** NS ** ** ** ** NS NS * CRE EC-Klf2-KO EC-Klf4-KO Klf2 Klf4 Nos3 Thbd ** 3 ** ** ** NS ** 3 3 NS NS ** NS NS NS CRE EC-Klf2-KO EC-Klf4-KO EC-DKO EC-DKO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO ** * * CRE EC-Klf2-KO EC-Klf4-KO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO EC-DKO CRE EC-Klf2-KO EC-Klf4-KO EC-DKO TM GAPDH enos GAPDH Supplemental Figure 1. Expression of Klf2, Klf4, and their targets in primary cardiac microvascular EC. Relative mrna expression levels of Klf2 (Kruppel-like family of transcription factor 2), Klf4, Nos3 (endothelial nitric oxide synthase), and Thbd (thrombomodulin) in primary cardiac microvascular EC isolated from mice at day 6 (A), day 5 (B), and day 4 (C) following tamoxifen (n=3-5 per genotype, each sample was pooled from 2 mice). (D) Representative western blot of primary cardiac microvascular EC using antibodies against KLF4, enos (endothelial nitric oxide synthase), TM (thrombomodulin), and GAPDH (endogenous glyceraldehyde 3-phosphate dehydrogenase). CRE: Cdh5(PAC)-Ert2cre, EC-DKO: EC-specific Klf2 and Klf4 double knockout, EC-Klf2-KO: EC-specific Klf2 knockout, EC-Klf4-KO: EC-specific Klf4 knockout. Sample in each group was pooled from 7-8 mice. *: P < 0.05, * *: P < 0.01, NS: Not significant. One-way ANOVA with Bonferroni's post hoc test. 2
3 Supplemental Figure Baseline Day 6 post-tamoxifen EKG (mv) CRE EKG (mv) EC-DKO ST EKG (mv) EC-Klf2-KO EKG (mv) EC-Klf4-KO ms Supplemental Figure 2. Electrocardiogram (EKG) recording. Representative EKG of CRE, EC-DKO, EC-Klf2-KO, and EC-Klf4-KO mice at baseline without tamoxifen (left) and day 6 post-tamoxifen injection (right). n=4-5 per genotype. CRE: Cdh5(PAC)-Ert2cre, EC-DKO: EC-specific Klf2 and Klf4 double knockout, EC-Klf2-KO: EC-specific Klf2 knockout, EC-Klf4-KO: EC-specific Klf4 knockout. 3
4 Supplemental Figure 3 A Cardiac function 60 NS ** NS 80 NS ** NS 40 NS NS p= ** ** FS (%) EF (%) CO (ml/min) CRE EC-Klf2-KO EC-Klf4-KO EC-DKO 0 CRE EC-Klf2-KO EC-Klf4-KO EC-DKO 0 CRE EC-Klf2-KO EC-Klf4-KO EC-DKO B 5 NS ** NS C 5 NS NS * Chamber size 5 NS ** NS LV/BW (mg/g) LVIDd (mm) LVIDs (mm) CRE EC-Klf2-KO EC-Klf4-KO EC-DKO 0 CRE EC-Klf2-KO EC-Klf4-KO EC-DKO 0 CRE EC-Klf2-KO EC-Klf4-KO EC-DKO Supplemental Figure 3. Echocardiographic analysis at day 6 after tamoxifen administration. (A) Fractional shortening (FS), ejection fraction (EF), and cardiac output (CO) in CRE (n=3), EC-Klf2-KO (4), EC-Klf4-KO (n=4), and EC-DKO mice (n=5). (B) Left ventricle (LV)-to-body weight (BW) ratio. (C) LV internal dimension at end diastole (LVIDd), and at end systole (LVIDs). CRE: Cdh5(PAC)-Ert2cre, EC-DKO: EC-specific Klf2 and Klf4 double knockout, EC-Klf2-KO: EC-specific Klf2 knockout, EC-Klf4-KO: EC-specific Klf4 knockout. Data are presented as mean ± SEM. *: P < 0.05, * *: P < NS: Not significant. One-way ANOVA with Bonferroni's post hoc test. 4
5 Supplemental Figure 4 EC-Klf2-KO EC-Klf4-KO Brain Lung Heart Skin Supplemental Figure 4. Gross post-mortem examination in EC-Klf2-KO and EC-Klf4-KO mice. Representative gross anatomy pictures of brain, lungs, heart, and subcutaneous tissues from EC-Klf2-KO (EC-specific Klf2 knockout) and EC-Klf4-KO (EC-specific Klf4 knockout) mice at day 6 post-tamoxifen (n=3-4 per genotype). 5
6 Supplemental Figure 5 A Eosin (Lung) EM (Lung) Evans blue (Lung) Evans blue (Kidney) CRE EC-DKO B Permeability (corrected E620) C Permeability (corrected E620) D Permeability (corrected E620) Day 5 post-tamoxifen Lung 0.5 Kidney 0.5 Heart ** 0.4 * Day 4 post-tamoxifen Lung Kidney Heart * 0.5 * 0.5 NS CRE EC-DKO CRE EC-DKO CRE EC-DKO CRE EC-DKO CRE EC-DKO CRE EC-DKO Day 6 post-tamoxifen Lung * Kidney * Heart CRE EC-DKO CRE EC-DKO CRE EC-DKO NS * Supplemental Figure 5. Endothelial-specific deletion of Klf2 and Klf4 results in vascular leak. (A, top) Representative pictures of lung Eosin staining show erythrocyte leakage (white arrow) in extravascular tissues at day 6 post-tamoxifen. (n=3-4 per genotype). Scale bar is 200 μm. (A, middle) Representative electron microscopic (EM) images of lung indicate discontinuity of endothelial monolayer at day 6 post-tamoxifen (n=3-4 per genotype). Scale bar is 2 μm. Endothelial cells (EC): red arrowhead, extravascular erythrocytes: white arrow, leucocytes: blue arrow. (A, bottom) Representative pictures of Evans blue vascular permeability assay for the lungs and kidneys at day 6 post-tamoxifen (n=7-8 per genotype). (B-D) Quantification of extravasated Evans Blue dye in the lungs, kidneys, and heart at day 4 (B, n=6-8 per genotype), day 5 (C, n=6-9 per genotype), and day 6 (D, n=7-8 per genotype) post-tamoxifen. CRE: Cdh5(PAC)-Ert2cre, EC-DKO: EC-specific Klf2 and Klf4 double knockout. Data are presented as mean ± SEM. *: P < 0.05, * *: P < 0.01, NS: Not significant. 2-tailed Student s t test. 6
7 Supplemental Figure 6 5 NS * ** CRE EC-DKO Relative mrna expression F3 Serpine1 F2rl3 Supplemental Figure 6. Expression of F3, Serpine1, and F2rl3 mrna in primary cardiac microvascular EC at day 6 post-tamoxifen. n=3-4 per genotype, each sample was pooled from 2 mice. F3: tissue factor, Serpine1: serine (or cysteine) peptidase inhibitor, clade E, member 1, F2rl3: coagulation factor II receptor-like 3, CRE: Cdh5(PAC)-Ert2cre, EC-DKO: EC-specific Klf2 and Klf4 double knockout. *: P < 0.05, * *: P < 0.01, NS: Not significant. 2-tailed Student s t test. 7
8 Supplemental Figure 7 EC-DKO EC-DKO EC-DKO EM (Brain) Supplemental Figure 7. Representative electron microscopic (EM) images of brain indicate degeneration of EC and extravascular erythrocytes in the EC-DKO mice at day 6 post-tamoxifen. Scale bar is 2 μm. Endothelial cells (EC): red arrowhead, extravascular erythrocytes: white arrow, leucocytes: blue arrow, vessel lumen: black arrowhead (n=3-4 per genotype). EC-DKO: EC-specific Klf2 and Klf4 double knockout. 8
9 Supplemental Table 1. Differentially expressed genes (q<0.05) in cardiac microvascular EC obtained from EC-DKO in comparison to CRE mice at day 6 post-tamoxifen. Ensembl_ID Gene Description EC-DKOvsCRE.log2FC EC-DKOvsCRE.RawP EC-DKOvsCRE.FDR ENSMUSG Gm7809 predicted gene E E-177 ENSMUSG Mest mesoderm specific transcript E E-109 ENSMUSG Ces2e carboxylesterase 2E E E-87 ENSMUSG Syt7 synaptotagmin VII E E-83 ENSMUSG Alpl alkaline phosphatase, liver/bone/kidney E E-77 ENSMUSG Apln apelin E E-70 ENSMUSG Casp3 caspase E E-65 ENSMUSG Tnc tenascin C E E-59 ENSMUSG Dpysl3 dihydropyrimidinase-like E E-58 ENSMUSG Ptprr protein tyrosine phosphatase, receptor type, R E E-57 ENSMUSG H2-T24 histocompatibility 2, T region locus E E-57 ENSMUSG Cd109 CD109 antigen E E-57 ENSMUSG Gm25360 predicted gene, E E-51 ENSMUSG Ceacam1 carcinoembryonic antigen-related cell adhesion molecule E E-51 ENSMUSG Prg4 proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) E E-50 ENSMUSG Sele selectin, endothelial cell E E-49 ENSMUSG Plau plasminogen activator, urokinase E E-46 ENSMUSG Tubb6 tubulin, beta 6 class V E E-45 ENSMUSG Ptchd1 patched domain containing E E-44 ENSMUSG Wisp1 WNT1 inducible signaling pathway protein E E-44 ENSMUSG Angpt2 angiopoietin E E-42 ENSMUSG Jam2 junction adhesion molecule E E-42 ENSMUSG Igf1 insulin-like growth factor E E-40 ENSMUSG Pcsk5 proprotein convertase subtilisin/kexin type E E-40 ENSMUSG Sparc secreted acidic cysteine rich glycoprotein E E-40 ENSMUSG Ahr aryl-hydrocarbon receptor E E-39 ENSMUSG Abcb4 ATP-binding cassette, sub-family B (MDR/TAP), member E E-39 ENSMUSG Slc30a4 solute carrier family 30 (zinc transporter), member E E-39 ENSMUSG Akr1c19 aldo-keto reductase family 1, member C E E-39 ENSMUSG Kcnj2 potassium inwardly-rectifying channel, subfamily J, member E E-39 ENSMUSG Nr3c2 nuclear receptor subfamily 3, group C, member E E-38 ENSMUSG Crispld1 cysteine-rich secretory protein LCCL domain containing E E-38 ENSMUSG Akr1b8 aldo-keto reductase family 1, member B E E-38 ENSMUSG Slc26a10 solute carrier family 26, member E E-37 ENSMUSG Prcp prolylcarboxypeptidase (angiotensinase C) E E-37 ENSMUSG Nuak1 NUAK family, SNF1-like kinase, E E-37 ENSMUSG Fam167b family with sequence similarity 167, member B E E-37 ENSMUSG Clca5 chloride channel calcium activated E E-36 ENSMUSG P05Rik RIKEN cdna P05 gene E E-36 ENSMUSG Vim vimentin E E-36 ENSMUSG Rassf9 Ras association (RalGDS/AF-6) domain family (N-terminal) member E E-36 ENSMUSG Tmsb10 thymosin, beta E E-35 ENSMUSG Tln2 talin E E-35 ENSMUSG Clmn calmin E E-35 ENSMUSG Tuba1c tubulin, alpha 1C E E-34 ENSMUSG Kank4 KN motif and ankyrin repeat domains E E-34 ENSMUSG Tll1 tolloid-like E E-33 ENSMUSG Myh10 myosin, heavy polypeptide 10, non-muscle E E-33 ENSMUSG Ces2g carboxylesterase 2G E E-33 ENSMUSG Rad51ap1 RAD51 associated protein E E-32 ENSMUSG Slc22a4 solute carrier family 22 (organic cation transporter), member E E-31 ENSMUSG Nrip2 nuclear receptor interacting protein E E-31 ENSMUSG Coro6 coronin E E-31 ENSMUSG Syt15 synaptotagmin XV E E-31 ENSMUSG Cxxc4 CXXC finger E E-31 ENSMUSG Aif1l allograft inflammatory factor 1-like E E-31 ENSMUSG Nepn nephrocan E E-30 ENSMUSG Map1b microtubule-associated protein 1B E E-30 ENSMUSG Nebl nebulette E E-29 ENSMUSG Pltp phospholipid transfer protein E E-29 ENSMUSG Cytl1 cytokine-like E E-29 ENSMUSG Acss1 acyl-coa synthetase short-chain family member E E-29 ENSMUSG Ankrd33b ankyrin repeat domain 33B E E-29 ENSMUSG Sqle squalene epoxidase E E-29 ENSMUSG Wnt9b wingless-type MMTV integration site family, member 9B E E-28 ENSMUSG Gm12216 predicted gene E E-28 ENSMUSG Slco1a4 solute carrier organic anion transporter family, member 1a E E-27 ENSMUSG Adamts17 a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, E E-27 ENSMUSG Dnm3 dynamin E E-27 ENSMUSG Cd276 CD276 antigen E E-27 ENSMUSG Magix MAGI family member, X-linked E E-27 Page 1 of 84
10 ENSMUSG Rasgrf2 RAS protein-specific guanine nucleotide-releasing factor E E-27 ENSMUSG Pitpnm3 PITPNM family member E E-27 ENSMUSG AU expressed sequence AU E E-27 ENSMUSG Gbp4 guanylate binding protein E E-27 ENSMUSG Pxdn peroxidasin homolog (Drosophila) E E-27 ENSMUSG Mamdc2 MAM domain containing E E-27 ENSMUSG Stmn2 stathmin-like E E-27 ENSMUSG Jazf1 JAZF zinc finger E E-26 ENSMUSG Slc28a2 solute carrier family 28 (sodium-coupled nucleoside transporter), member E E-26 ENSMUSG Thbd thrombomodulin E E-26 ENSMUSG Wbp5 WW domain binding protein E E-26 ENSMUSG Prrt4 proline-rich transmembrane protein E E-26 ENSMUSG Sidt1 SID1 transmembrane family, member E E-26 ENSMUSG Sybu syntabulin (syntaxin-interacting) E E-26 ENSMUSG Fbln5 fibulin E E-26 ENSMUSG E2f1 E2F transcription factor E E-26 ENSMUSG Rhpn2 rhophilin, Rho GTPase binding protein E E-26 ENSMUSG Cdkn1c cyclin-dependent kinase inhibitor 1C (P57) E E-26 ENSMUSG Lpar4 lysophosphatidic acid receptor E E-26 ENSMUSG S100a11 S100 calcium binding protein A11 (calgizzarin) E E-26 ENSMUSG Litaf LPS-induced TN factor E E-26 ENSMUSG Sgpp2 sphingosine-1-phosphate phosphotase E E-25 ENSMUSG Tmtc1 transmembrane and tetratricopeptide repeat containing E E-25 ENSMUSG Scn4b sodium channel, type IV, beta E E-25 ENSMUSG Dll1 delta-like 1 (Drosophila) E E-25 ENSMUSG Myof myoferlin E E-25 ENSMUSG Entpd1 ectonucleoside triphosphate diphosphohydrolase E E-25 ENSMUSG Txlnb taxilin beta E E-25 ENSMUSG Fstl1 follistatin-like E E-25 ENSMUSG Fndc5 fibronectin type III domain containing E E-25 ENSMUSG Mn1 meningioma E E-24 ENSMUSG Prom1 prominin E E-24 ENSMUSG Rell1 RELT-like E E-24 ENSMUSG Anxa1 annexin A E E-24 ENSMUSG Nid1 nidogen E E-24 ENSMUSG Arl4d ADP-ribosylation factor-like 4D E E-24 ENSMUSG Sdk1 sidekick homolog 1 (chicken) E E-24 ENSMUSG Acacb acetyl-coenzyme A carboxylase beta E E-24 ENSMUSG Gm14290 predicted gene E E-24 ENSMUSG Podxl podocalyxin-like E E-23 ENSMUSG Etv4 ets variant E E-23 ENSMUSG Sh2d5 SH2 domain containing E E-23 ENSMUSG Acan aggrecan E E-23 ENSMUSG Ywhah tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide E E-23 ENSMUSG Disp2 dispatched homolog 2 (Drosophila) E E-23 ENSMUSG Zfp185 zinc finger protein E E-23 ENSMUSG Cdc25c cell division cycle 25C E E-23 ENSMUSG Car13 carbonic anhydrase E E-23 ENSMUSG Mtmr11 myotubularin related protein E E-22 ENSMUSG Ccnb2 cyclin B E E-22 ENSMUSG Man1a mannosidase 1, alpha E E-22 ENSMUSG Mettl7a1 methyltransferase like 7A E E-22 ENSMUSG B4galt3 UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide E E-22 ENSMUSG Slc6a6 solute carrier family 6 (neurotransmitter transporter, taurine), member E E-22 ENSMUSG Pgk1 phosphoglycerate kinase E E-22 ENSMUSG Kit kit oncogene E E-22 ENSMUSG Spice1 spindle and centriole associated protein E E-21 ENSMUSG Bmp2 bone morphogenetic protein E E-21 ENSMUSG Mbnl3 muscleblind-like 3 (Drosophila) E E-21 ENSMUSG Kifc2 kinesin family member C E E-21 ENSMUSG Npnt nephronectin E E-21 ENSMUSG Tmem56 transmembrane protein E E-21 ENSMUSG Atp1a2 ATPase, Na+/K+ transporting, alpha 2 polypeptide E E-21 ENSMUSG Lamb1 laminin B E E-21 ENSMUSG Dhrs3 dehydrogenase/reductase (SDR family) member E E-21 ENSMUSG Myoz2 myozenin E E-21 ENSMUSG Fkbp1a FK506 binding protein 1a E E-21 ENSMUSG Kcnk3 potassium channel, subfamily K, member E E-21 ENSMUSG Col13a1 collagen, type XIII, alpha E E-21 ENSMUSG Sh3d21 SH3 domain containing E E-21 ENSMUSG Nxpe2 neurexophilin and PC-esterase domain family, member E E-21 ENSMUSG St6galnac2 ST6 (alpha-n-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-n-acetylgalactosaminide alpha-2,6-sialyltransferase E E-21 ENSMUSG Hdac11 histone deacetylase E E-21 Page 2 of 84
11 ENSMUSG Sh3pxd2b SH3 and PX domains 2B E E-21 ENSMUSG Gm16070 predicted gene E E-21 ENSMUSG Mrgprh MAS-related GPR, member H E E-21 ENSMUSG Morf4l2 mortality factor 4 like E E-20 ENSMUSG Adamtsl1 ADAMTS-like E E-20 ENSMUSG Pbld1 phenazine biosynthesis-like protein domain containing E E-20 ENSMUSG Rassf2 Ras association (RalGDS/AF-6) domain family member E E-20 ENSMUSG Sacs sacsin E E-20 ENSMUSG Cd93 CD93 antigen E E-20 ENSMUSG Slc38a11 solute carrier family 38, member E E-20 ENSMUSG Gbp5 guanylate binding protein E E-20 ENSMUSG Nus1 nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) E E-20 ENSMUSG Arhgap11a Rho GTPase activating protein 11A E E-20 ENSMUSG Arf2 ADP-ribosylation factor E E-20 ENSMUSG Pde4b phosphodiesterase 4B, camp specific E E-20 ENSMUSG Vegfc vascular endothelial growth factor C E E-20 ENSMUSG Slc12a7 solute carrier family 12, member E E-19 ENSMUSG Plp2 proteolipid protein E E-19 ENSMUSG Nts neurotensin E E-19 ENSMUSG Kazald1 Kazal-type serine peptidase inhibitor domain E E-19 ENSMUSG Sgms2 sphingomyelin synthase E E-19 ENSMUSG Mfsd7c major facilitator superfamily domain containing 7C E E-19 ENSMUSG Ulbp1 UL16 binding protein E E-19 ENSMUSG Pvr poliovirus receptor E E-19 ENSMUSG Cyp27a1 cytochrome P450, family 27, subfamily a, polypeptide E E-19 ENSMUSG Prnp prion protein E E-19 ENSMUSG Pygm muscle glycogen phosphorylase E E-18 ENSMUSG Rtn4 reticulon E E-18 ENSMUSG Ppa1 pyrophosphatase (inorganic) E E-18 ENSMUSG Plk1 polo-like kinase E E-18 ENSMUSG Itga2 integrin alpha E E-18 ENSMUSG Arhgef26 Rho guanine nucleotide exchange factor (GEF) E E-18 ENSMUSG Chst1 carbohydrate (keratan sulfate Gal-6) sulfotransferase E E-18 ENSMUSG Zdhhc15 zinc finger, DHHC domain containing E E-18 ENSMUSG J21Rik1 RIKEN cdna J21 gene E E-18 ENSMUSG A02Rik RIKEN cdna A02 gene E E-18 ENSMUSG Wscd1 WSC domain containing E E-18 ENSMUSG Ckap4 cytoskeleton-associated protein E E-18 ENSMUSG Cks1b CDC28 protein kinase 1b E E-18 ENSMUSG Hist1h1d histone cluster 1, H1d E E-18 ENSMUSG Kdelr3 KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor E E-18 ENSMUSG Robo2 roundabout homolog 2 (Drosophila) E E-18 ENSMUSG Lgals1 lectin, galactose binding, soluble E E-17 ENSMUSG Tmem47 transmembrane protein E E-17 ENSMUSG Lims1 LIM and senescent cell antigen-like domains E E-17 ENSMUSG Kcne3 potassium voltage-gated channel, Isk-related subfamily, gene E E-17 ENSMUSG F2rl3 coagulation factor II (thrombin) receptor-like E E-17 ENSMUSG Pdzd2 PDZ domain containing E E-17 ENSMUSG Als2cl ALS2 C-terminal like E E-17 ENSMUSG Fn3k fructosamine 3 kinase E E-17 ENSMUSG Tacc3 transforming, acidic coiled-coil containing protein E E-17 ENSMUSG Ppic peptidylprolyl isomerase C E E-17 ENSMUSG Gper1 G protein-coupled estrogen receptor E E-17 ENSMUSG Exoc3l4 exocyst complex component 3-like E E-17 ENSMUSG Mcam melanoma cell adhesion molecule E E-17 ENSMUSG Smad9 SMAD family member E E-17 ENSMUSG Gtse1 G two S phase expressed protein E E-17 ENSMUSG Pfn1 profilin E E-17 ENSMUSG Suv39h2 suppressor of variegation 3-9 homolog 2 (Drosophila) E E-17 ENSMUSG Arsi arylsulfatase i E E-17 ENSMUSG Prrg3 proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) E E-17 ENSMUSG Stard8 START domain containing E E-17 ENSMUSG Vcan versican E E-17 ENSMUSG Galnt1 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase E E-17 ENSMUSG Klk8 kallikrein related-peptidase E E-17 ENSMUSG Marcksl1 MARCKS-like E E-17 ENSMUSG P07Rik RIKEN cdna P07 gene E E-17 ENSMUSG Lyrm9 LYR motif containing E E-17 ENSMUSG Nid2 nidogen E E-17 ENSMUSG Hrh2 histamine receptor H E E-17 ENSMUSG Csrp1 cysteine and glycine-rich protein E E-17 ENSMUSG Scamp5 secretory carrier membrane protein E E-17 ENSMUSG Pvrl2 poliovirus receptor-related E E-17 Page 3 of 84
12 ENSMUSG Ube2c ubiquitin-conjugating enzyme E2C E E-17 ENSMUSG Cyp1a1 cytochrome P450, family 1, subfamily a, polypeptide E E-17 ENSMUSG Gng2 guanine nucleotide binding protein (G protein), gamma E E-17 ENSMUSG Dgkb diacylglycerol kinase, beta E E-17 ENSMUSG Ighm immunoglobulin heavy constant mu E E-16 ENSMUSG Rassf10 Ras association (RalGDS/AF-6) domain family (N-terminal) member E E-16 ENSMUSG Tmem255a transmembrane protein 255A E E-16 ENSMUSG Aqp7 aquaporin E E-16 ENSMUSG Tnfrsf1b tumor necrosis factor receptor superfamily, member 1b E E-16 ENSMUSG Dclk1 doublecortin-like kinase E E-16 ENSMUSG Uhrf1bp1l UHRF1 (ICBP90) binding protein 1-like E E-16 ENSMUSG Adra2c adrenergic receptor, alpha 2c E E-16 ENSMUSG Dsp desmoplakin E E-16 ENSMUSG Gm694 predicted gene E E-16 ENSMUSG Ntng2 netrin G E E-16 ENSMUSG Gpsm2 G-protein signalling modulator 2 (AGS3-like, C. elegans) E E-16 ENSMUSG Upp1 uridine phosphorylase E E-16 ENSMUSG Cxcl12 chemokine (C-X-C motif) ligand E E-16 ENSMUSG Vmp1 vacuole membrane protein E E-16 ENSMUSG Gpr157 G protein-coupled receptor E E-16 ENSMUSG O22Rik RIKEN cdna O22 gene E E-16 ENSMUSG Epha4 Eph receptor A E E-16 ENSMUSG Fmo5 flavin containing monooxygenase E E-16 ENSMUSG Tmie transmembrane inner ear E E-16 ENSMUSG Prickle1 prickle homolog 1 (Drosophila) E E-16 ENSMUSG Cd200 CD200 antigen E E-16 ENSMUSG Tgtp1 T cell specific GTPase E E-16 ENSMUSG Cd44 CD44 antigen E E-16 ENSMUSG Pdk2 pyruvate dehydrogenase kinase, isoenzyme E E-16 ENSMUSG Rasgrp2 RAS, guanyl releasing protein E E-16 ENSMUSG Nes nestin E E-16 ENSMUSG Tnfrsf9 tumor necrosis factor receptor superfamily, member E E-16 ENSMUSG Stra6 stimulated by retinoic acid gene E E-16 ENSMUSG Procr protein C receptor, endothelial E E-16 ENSMUSG Slfn2 schlafen E E-16 ENSMUSG Eef1a2 eukaryotic translation elongation factor 1 alpha E E-16 ENSMUSG Car7 carbonic anhydrase E E-16 ENSMUSG Itgb1 integrin beta 1 (fibronectin receptor beta) E E-16 ENSMUSG Nod2 nucleotide-binding oligomerization domain containing E E-16 ENSMUSG Snn stannin E E-16 ENSMUSG Zfp52 zinc finger protein E E-16 ENSMUSG Tspan2 tetraspanin E E-16 ENSMUSG Aldh5a1 aldhehyde dehydrogenase family 5, subfamily A E E-16 ENSMUSG Cd274 CD274 antigen E E-16 ENSMUSG Smpdl3a sphingomyelin phosphodiesterase, acid-like 3A E E-16 ENSMUSG Sdcbp syndecan binding protein E E-16 ENSMUSG Akap6 A kinase (PRKA) anchor protein E E-16 ENSMUSG Cenpt centromere protein T E E-16 ENSMUSG Prr5l proline rich 5 like E E-16 ENSMUSG Pdgfa platelet derived growth factor, alpha E E-16 ENSMUSG Tagln2 transgelin E E-16 ENSMUSG Hist1h3b histone cluster 1, H3b E E-16 ENSMUSG Calu calumenin E E-16 ENSMUSG Pfkp phosphofructokinase, platelet E E-16 ENSMUSG Trpv1 transient receptor potential cation channel, subfamily V, member E E-15 ENSMUSG Kcnb1 potassium voltage gated channel, Shab-related subfamily, member E E-15 ENSMUSG Incenp inner centromere protein E E-15 ENSMUSG Rundc3b RUN domain containing 3B E E-15 ENSMUSG Prob1 proline rich basic protein E E-15 ENSMUSG Ablim2 actin-binding LIM protein E E-15 ENSMUSG Ppp1r12b protein phosphatase 1, regulatory (inhibitor) subunit 12B E E-15 ENSMUSG Myl2 myosin, light polypeptide 2, regulatory, cardiac, slow E E-15 ENSMUSG Nek6 NIMA (never in mitosis gene a)-related expressed kinase E E-15 ENSMUSG Myrip myosin VIIA and Rab interacting protein E E-15 ENSMUSG Mapk6 mitogen-activated protein kinase E E-15 ENSMUSG Olfr1396 olfactory receptor E E-15 ENSMUSG Cyp2d22 cytochrome P450, family 2, subfamily d, polypeptide E E-15 ENSMUSG Cyfip2 cytoplasmic FMR1 interacting protein E E-15 ENSMUSG Lif leukemia inhibitory factor E E-15 ENSMUSG Sdf2l1 stromal cell-derived factor 2-like E E-15 ENSMUSG L13Rik RIKEN cdna L13 gene E E-15 ENSMUSG Rad18 RAD18 homolog (S. cerevisiae) E E-15 ENSMUSG Il2ra interleukin 2 receptor, alpha chain E E-15 Page 4 of 84
13 ENSMUSG Rhbdl2 rhomboid, veinlet-like 2 (Drosophila) E E-15 ENSMUSG Ammecr1 Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene E E-15 ENSMUSG Ntf3 neurotrophin E E-15 ENSMUSG Csgalnact1 chondroitin sulfate N-acetylgalactosaminyltransferase E E-15 ENSMUSG Lrp8 low density lipoprotein receptor-related protein 8, apolipoprotein e receptor E E-15 ENSMUSG Emp1 epithelial membrane protein E E-15 ENSMUSG Lrrc59 leucine rich repeat containing E E-15 ENSMUSG Cbx7 chromobox E E-15 ENSMUSG Alox12 arachidonate 12-lipoxygenase E E-15 ENSMUSG Mrvi1 MRV integration site E E-15 ENSMUSG F22Rik RIKEN cdna F22 gene E E-15 ENSMUSG H2-Ob histocompatibility 2, O region beta locus E E-15 ENSMUSG Hist1h1a histone cluster 1, H1a E E-15 ENSMUSG Fam83d family with sequence similarity 83, member D E E-15 ENSMUSG Gpr20 G protein-coupled receptor E E-15 ENSMUSG M07Rik RIKEN cdna M07 gene E E-15 ENSMUSG Ripk3 receptor-interacting serine-threonine kinase E E-15 ENSMUSG Lamc1 laminin, gamma E E-14 ENSMUSG Cdk1 cyclin-dependent kinase E E-14 ENSMUSG Armc2 armadillo repeat containing E E-14 ENSMUSG Pdlim1 PDZ and LIM domain 1 (elfin) E E-14 ENSMUSG Prelp proline arginine-rich end leucine-rich repeat E E-14 ENSMUSG Bambi-ps1 BMP and activin membrane-bound inhibitor, pseudogene (Xenopus laevis) E E-14 ENSMUSG Runx1t1 runt-related transcription factor 1; translocated to, 1 (cyclin D-related) E E-14 ENSMUSG Cdc20 cell division cycle E E-14 ENSMUSG Ctla2a cytotoxic T lymphocyte-associated protein 2 alpha E E-14 ENSMUSG Eno3 enolase 3, beta muscle E E-14 ENSMUSG Nov nephroblastoma overexpressed gene E E-14 ENSMUSG Kif2c kinesin family member 2C E E-14 ENSMUSG Ly6c1 lymphocyte antigen 6 complex, locus C E E-14 ENSMUSG Nek2 NIMA (never in mitosis gene a)-related expressed kinase E E-14 ENSMUSG Pisd phosphatidylserine decarboxylase E E-14 ENSMUSG Atp10d ATPase, class V, type 10D E E-14 ENSMUSG Syne1 spectrin repeat containing, nuclear envelope E E-14 ENSMUSG Cmpk1 cytidine monophosphate (UMP-CMP) kinase E E-14 ENSMUSG Trim65 tripartite motif-containing E E-14 ENSMUSG Pdia6 protein disulfide isomerase associated E E-14 ENSMUSG Lama3 laminin, alpha E E-14 ENSMUSG Slc25a23 solute carrier family 25 (mitochondrial carrier; phosphate carrier), member E E-14 ENSMUSG Slc2a4 solute carrier family 2 (facilitated glucose transporter), member E E-14 ENSMUSG Spred3 sprouty-related, EVH1 domain containing E E-14 ENSMUSG Ptprg protein tyrosine phosphatase, receptor type, G E E-14 ENSMUSG Clic1 chloride intracellular channel E E-14 ENSMUSG Kifc1 kinesin family member C E E-14 ENSMUSG Tcea3 transcription elongation factor A (SII), E E-14 ENSMUSG Edem1 ER degradation enhancer, mannosidase alpha-like E E-14 ENSMUSG Troap trophinin associated protein E E-14 ENSMUSG Irf8 interferon regulatory factor E E-14 ENSMUSG Ctla2b cytotoxic T lymphocyte-associated protein 2 beta E E-14 ENSMUSG Kif22 kinesin family member E E-14 ENSMUSG Flrt2 fibronectin leucine rich transmembrane protein E E-14 ENSMUSG Aspa aspartoacylase E E-14 ENSMUSG Pnkd paroxysmal nonkinesiogenic dyskinesia E E-14 ENSMUSG Bean1 brain expressed, associated with Nedd4, E E-14 ENSMUSG Sun1 Sad1 and UNC84 domain containing E E-14 ENSMUSG Kcnmb4 potassium large conductance calcium-activated channel, subfamily M, beta member E E-14 ENSMUSG Ret ret proto-oncogene E E-14 ENSMUSG Tuba1a tubulin, alpha 1A E E-14 ENSMUSG Col5a2 collagen, type V, alpha E E-14 ENSMUSG Fam78b family with sequence similarity 78, member B E E-14 ENSMUSG Tex2 testis expressed gene E E-14 ENSMUSG Synj2 synaptojanin E E-14 ENSMUSG Psmd14 proteasome (prosome, macropain) 26S subunit, non-atpase, E E-13 ENSMUSG Vash1 vasohibin E E-13 ENSMUSG Hcls1 hematopoietic cell specific Lyn substrate E E-13 ENSMUSG Arrdc3 arrestin domain containing E E-13 ENSMUSG Stil Scl/Tal1 interrupting locus E E-13 ENSMUSG Cfl1 cofilin 1, non-muscle E E-13 ENSMUSG Spats2 spermatogenesis associated, serine-rich E E-13 ENSMUSG Cxcl10 chemokine (C-X-C motif) ligand E E-13 ENSMUSG Samsn1 SAM domain, SH3 domain and nuclear localization signals, E E-13 ENSMUSG Pno1 partner of NOB1 homolog (S. cerevisiae) E E-13 ENSMUSG Manf mesencephalic astrocyte-derived neurotrophic factor E E-13 Page 5 of 84
14 ENSMUSG Grip2 glutamate receptor interacting protein E E-13 ENSMUSG Rab6b RAB6B, member RAS oncogene family E E-13 ENSMUSG Il1a interleukin 1 alpha E E-13 ENSMUSG Bora bora, aurora kinase A activator E E-13 ENSMUSG Lxn latexin E E-13 ENSMUSG Tmem38a transmembrane protein 38A E E-13 ENSMUSG Hif1a hypoxia inducible factor 1, alpha subunit E E-13 ENSMUSG Ctsf cathepsin F E E-13 ENSMUSG Kcnn3 potassium intermediate/small conductance calcium-activated channel, subfamily N, member E E-13 ENSMUSG Hn1l hematological and neurological expressed 1-like E E-13 ENSMUSG Plce1 phospholipase C, epsilon E E-13 ENSMUSG Cacna1g calcium channel, voltage-dependent, T type, alpha 1G subunit E E-13 ENSMUSG Atp7a ATPase, Cu++ transporting, alpha polypeptide E E-13 ENSMUSG Ppfibp2 PTPRF interacting protein, binding protein 2 (liprin beta 2) E E-13 ENSMUSG Tdpx-ps1 thioredoxin peroxidase, pseudogene 1 Inf 3.74E E-13 ENSMUSG Akap7 A kinase (PRKA) anchor protein E E-13 ENSMUSG Psmd8 proteasome (prosome, macropain) 26S subunit, non-atpase, E E-13 ENSMUSG Gm9917 predicted gene E E-13 ENSMUSG Bmp2k BMP2 inducible kinase E E-13 ENSMUSG Adamtsl3 ADAMTS-like E E-13 ENSMUSG Myadm myeloid-associated differentiation marker E E-13 ENSMUSG Ppm1l protein phosphatase 1 (formerly 2C)-like E E-13 ENSMUSG Tspan18 tetraspanin E E-13 ENSMUSG Soat1 sterol O-acyltransferase E E-13 ENSMUSG AI expressed sequence AI E E-13 ENSMUSG Anxa2 annexin A E E-13 ENSMUSG Gm22973 predicted gene, E E-13 ENSMUSG Trpc3 transient receptor potential cation channel, subfamily C, member E E-13 ENSMUSG Sned1 sushi, nidogen and EGF-like domains E E-13 ENSMUSG Cnksr3 Cnksr family member E E-13 ENSMUSG Fam49a family with sequence similarity 49, member A E E-13 ENSMUSG Cyyr1 cysteine and tyrosine-rich protein E E-13 ENSMUSG Crhbp corticotropin releasing hormone binding protein E E-13 ENSMUSG Cdca8 cell division cycle associated E E-13 ENSMUSG Phf1 PHD finger protein E E-13 ENSMUSG Tmc5 transmembrane channel-like gene family E E-13 ENSMUSG Acsl4 acyl-coa synthetase long-chain family member E E-13 ENSMUSG Col4a1 collagen, type IV, alpha E E-13 ENSMUSG Cst3 cystatin C E E-13 ENSMUSG Pkig protein kinase inhibitor, gamma E E-13 ENSMUSG Kif20a kinesin family member 20A E E-13 ENSMUSG Acot7 acyl-coa thioesterase E E-13 ENSMUSG Klk10 kallikrein related-peptidase E E-13 ENSMUSG Timp1 tissue inhibitor of metalloproteinase E E-13 ENSMUSG Prkg2 protein kinase, cgmp-dependent, type II E E-13 ENSMUSG Wdr62 WD repeat domain E E-13 ENSMUSG Nudt12 nudix (nucleoside diphosphate linked moiety X)-type motif E E-13 ENSMUSG Mlkl mixed lineage kinase domain-like E E-13 ENSMUSG Tnfrsf23 tumor necrosis factor receptor superfamily, member E E-13 ENSMUSG Oas2 2'-5' oligoadenylate synthetase E E-13 ENSMUSG Knstrn kinetochore-localized astrin/spag5 binding E E-13 ENSMUSG Psmc4 proteasome (prosome, macropain) 26S subunit, ATPase, E E-13 ENSMUSG Rnf122 ring finger protein E E-13 ENSMUSG Fam117a family with sequence similarity 117, member A E E-13 ENSMUSG Fhl3 four and a half LIM domains E E-13 ENSMUSG Slc39a6 solute carrier family 39 (metal ion transporter), member E E-13 ENSMUSG C19Rik RIKEN cdna C19 gene E E-13 ENSMUSG Iqgap3 IQ motif containing GTPase activating protein E E-13 ENSMUSG Myh14 myosin, heavy polypeptide E E-13 ENSMUSG Slfn3 schlafen E E-13 ENSMUSG Gucy1a3 guanylate cyclase 1, soluble, alpha E E-13 ENSMUSG Gars glycyl-trna synthetase E E-13 ENSMUSG Pcsk6 proprotein convertase subtilisin/kexin type E E-12 ENSMUSG Mmp14 matrix metallopeptidase 14 (membrane-inserted) E E-12 ENSMUSG Mill2 MHC I like leukocyte E E-12 ENSMUSG Ddias DNA damage-induced apoptosis suppressor E E-12 ENSMUSG Nxpe4 neurexophilin and PC-esterase domain family, member E E-12 ENSMUSG Ppap2c phosphatidic acid phosphatase type 2C E E-12 ENSMUSG Fam167a family with sequence similarity 167, member A E E-12 ENSMUSG Rhoc ras homolog gene family, member C E E-12 ENSMUSG Actr3 ARP3 actin-related protein E E-12 ENSMUSG P21Rik RIKEN cdna P21 gene E E-12 ENSMUSG Pln phospholamban E E-12 Page 6 of 84
15 ENSMUSG Zmat3 zinc finger matrin type E E-12 ENSMUSG Tnfaip8l1 tumor necrosis factor, alpha-induced protein 8-like E E-12 ENSMUSG Erbb3 v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) E E-12 ENSMUSG Garem GRB2 associated, regulator of MAPK E E-12 ENSMUSG Cenpq centromere protein Q E E-12 ENSMUSG Arpc5 actin related protein 2/3 complex, subunit E E-12 ENSMUSG Tnfsf10 tumor necrosis factor (ligand) superfamily, member E E-12 ENSMUSG Tcf23 transcription factor E E-12 ENSMUSG Memo1 mediator of cell motility E E-12 ENSMUSG Cyp51 cytochrome P450, family E E-12 ENSMUSG Sema3a sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A E E-12 ENSMUSG C1qtnf6 C1q and tumor necrosis factor related protein E E-12 ENSMUSG Synpo2 synaptopodin E E-12 ENSMUSG Cd22 CD22 antigen E E-12 ENSMUSG Fam20a family with sequence similarity 20, member A E E-12 ENSMUSG Nrbp2 nuclear receptor binding protein E E-12 ENSMUSG Dgat2 diacylglycerol O-acyltransferase E E-12 ENSMUSG Wfdc1 WAP four-disulfide core domain E E-12 ENSMUSG Fgfr3 fibroblast growth factor receptor E E-12 ENSMUSG Nin ninein E E-12 ENSMUSG Svip small VCP/p97-interacting protein E E-12 ENSMUSG Asb2 ankyrin repeat and SOCS box-containing E E-12 ENSMUSG Zrsr1 zinc finger (CCCH type), RNA binding motif and serine/arginine rich E E-12 ENSMUSG Ldlr low density lipoprotein receptor E E-12 ENSMUSG Mtus2 microtubule associated tumor suppressor candidate E E-12 ENSMUSG Mgat5 mannoside acetylglucosaminyltransferase E E-12 ENSMUSG Asns asparagine synthetase E E-12 ENSMUSG Thrb thyroid hormone receptor beta E E-12 ENSMUSG Nudt4 nudix (nucleoside diphosphate linked moiety X)-type motif E E-12 ENSMUSG Dnase2a deoxyribonuclease II alpha E E-12 ENSMUSG Thyn1 thymocyte nuclear protein E E-12 ENSMUSG Lyrm5 LYR motif containing E E-12 ENSMUSG Igfbp7 insulin-like growth factor binding protein E E-12 ENSMUSG Ip6k3 inositol hexaphosphate kinase E E-12 ENSMUSG Chd7 chromodomain helicase DNA binding protein E E-12 ENSMUSG Hist1h1b histone cluster 1, H1b E E-12 ENSMUSG Cmah cytidine monophospho-n-acetylneuraminic acid hydroxylase E E-12 ENSMUSG Txn1 thioredoxin E E-12 ENSMUSG Pif1 PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) E E-12 ENSMUSG Gm13861 predicted gene E E-12 ENSMUSG Ecm2 extracellular matrix protein 2, female organ and adipocyte specific E E-12 ENSMUSG Itga9 integrin alpha E E-12 ENSMUSG Fabp5 fatty acid binding protein 5, epidermal E E-12 ENSMUSG Cd40 CD40 antigen E E-12 ENSMUSG Naa50 N(alpha)-acetyltransferase 50, NatE catalytic subunit E E-12 ENSMUSG Calr3 calreticulin E E-12 ENSMUSG Tmem140 transmembrane protein E E-12 ENSMUSG Ephx1 epoxide hydrolase 1, microsomal E E-12 ENSMUSG RP24-422C E E-12 ENSMUSG Cda cytidine deaminase E E-12 ENSMUSG Plcb1 phospholipase C, beta E E-12 ENSMUSG Tada1 transcriptional adaptor E E-12 ENSMUSG Dnah6 dynein, axonemal, heavy chain E E-12 ENSMUSG AA expressed sequence AA E E-12 ENSMUSG Smad1 SMAD family member E E-12 ENSMUSG Bmx BMX non-receptor tyrosine kinase E E-12 ENSMUSG Asb10 ankyrin repeat and SOCS box-containing E E-12 ENSMUSG Ncaph non-smc condensin I complex, subunit H E E-12 ENSMUSG Abracl ABRA C-terminal like E E-12 ENSMUSG Nipal3 NIPA-like domain containing E E-12 ENSMUSG Klhl38 kelch-like E E-12 ENSMUSG Aurka aurora kinase A E E-12 ENSMUSG Asb18 ankyrin repeat and SOCS box-containing E E-12 ENSMUSG Kif18b kinesin family member 18B E E-12 ENSMUSG Tubb2b tubulin, beta 2B class IIB E E-11 ENSMUSG Pgm2l1 phosphoglucomutase 2-like E E-11 ENSMUSG Crmp1 collapsin response mediator protein E E-11 ENSMUSG Sstr4 somatostatin receptor E E-11 ENSMUSG Setbp1 SET binding protein E E-11 ENSMUSG Olfr78 olfactory receptor E E-11 ENSMUSG Hspa5 heat shock protein E E-11 ENSMUSG Mgat4a mannoside acetylglucosaminyltransferase 4, isoenzyme A E E-11 ENSMUSG Gm16340 predicted gene E E-11 Page 7 of 84
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ ΩΝ ΚΑΤΑ ΤΗΝ ΕΜΒΡΥΪΚΗ ΑΝΑΠΤΥΞΗ
 ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
Legends for Supplemental Data
 Legends for Supplemental Data Supplemental Fig. 1. Top molecular and cellular functions identified by Ingenuity Pathway Analysis after 6 h (left) and 24 h (right) of exposure of mouse pancreatic islets
Legends for Supplemental Data Supplemental Fig. 1. Top molecular and cellular functions identified by Ingenuity Pathway Analysis after 6 h (left) and 24 h (right) of exposure of mouse pancreatic islets
Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ ΡΑΣΗ ΤΟΥ ΓΑΣΤΡΙΚΟΥ ΒΛΕΝΝΟΓΟΝΟΥ ΣΤΗ ΛΟΙΜΩΞΗ ΜΕ ΕΛΙΚΟΒΑΚΤΗΡΙ ΙΟ ΤΟΥ ΠΥΛΩΡΟΥ ΠΡΙΝ ΚΑΙ ΜΕΤΑ ΤΗ ΘΕΡΑΠΕΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
ΙΩΑΝΝΗΣ ΚΛΑΓΚΑΣ. Χημικός, MSc ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ ΥΠΟΒΛΗΘΗΚΕ ΣΤΗΝ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΥ ΑΡΙΣΤΟΤΕΛΕΙΟΥ ΠΑΝΕΠΙΣΤΗΜΙΟΥ ΘΕΣΣΑΛΟΝΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΦΥΣΙΟΛΟΓΙΑΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ B ΕΡΓΑΣΤΗΡΙΟ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΔΙΕΥΘΥΝΤΗΣ: ΚΑΘΗΓΗΤΗΣ ΓΙΩΡΓΟΣ ΚΑΡΑΚΙΟΥΛΑΚΗΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 ΑΡΙΘΜ. 2376 Η ΕΠΙΔΡΑΣΗ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΦΥΣΙΟΛΟΓΙΑΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ B ΕΡΓΑΣΤΗΡΙΟ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΔΙΕΥΘΥΝΤΗΣ: ΚΑΘΗΓΗΤΗΣ ΓΙΩΡΓΟΣ ΚΑΡΑΚΙΟΥΛΑΚΗΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 ΑΡΙΘΜ. 2376 Η ΕΠΙΔΡΑΣΗ
Debashish Sahay. To cite this version: HAL Id: tel
 Identification of genes activated and biological markers involved in lysophosphatidic acid (LPA)-induced breast cancer metastasis through its receptor LPA1 Debashish Sahay To cite this version: Debashish
Identification of genes activated and biological markers involved in lysophosphatidic acid (LPA)-induced breast cancer metastasis through its receptor LPA1 Debashish Sahay To cite this version: Debashish
ΥΠΕΡΙΝΣΟΥΛΙΝΑΙΜΙΑ ΚΑΙ ΑΝΤΙΣΤΑΣΗ ΣΤΗΝ ΙΝΣΟΥΛΙΝΗ ΣΕ ΠΑΧΥΣΑΡΚΑ ΠΑΙΔΙΑ ΚΑΙ ΕΦΗΒΟΥΣ ΜΕ ΠΡΩΙΜΗ ΑΔΡΕΝΑΡΧΗ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΒΙΟΛΟΓΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΠΡΟΛΗΠΤΙΚΗΣ ΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΒΙΟΧΗΜΕΙΑΣ ΔΙΕΥΘΥΝΤΡΙΑ: Η ΚΑΘΗΓΗΤΡΙΑ Ν. ΒΑΒΑΤΣΗ-ΧΡΙΣΤΑΚΗ ΠΑΝΕΠ. ΕΤΟΣ 2009-2010 Αριθμ.2456
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΒΙΟΛΟΓΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΠΡΟΛΗΠΤΙΚΗΣ ΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΒΙΟΧΗΜΕΙΑΣ ΔΙΕΥΘΥΝΤΡΙΑ: Η ΚΑΘΗΓΗΤΡΙΑ Ν. ΒΑΒΑΤΣΗ-ΧΡΙΣΤΑΚΗ ΠΑΝΕΠ. ΕΤΟΣ 2009-2010 Αριθμ.2456
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Hvordan påvirker CO 2 post-smolt i brakkvann RAS?
 Hvordan påvirker CO 2 post-smolt i brakkvann RAS? Tom O. Nilsen,. Mota, V.C., Kolarevic, J., Ytterborg, E., Ebbesson, Lars O.E., Handeland, S.O., Stiller, K., Terjesen, B.F SUNNDALSØRA, 12. NOVEMBER 2018
Hvordan påvirker CO 2 post-smolt i brakkvann RAS? Tom O. Nilsen,. Mota, V.C., Kolarevic, J., Ytterborg, E., Ebbesson, Lars O.E., Handeland, S.O., Stiller, K., Terjesen, B.F SUNNDALSØRA, 12. NOVEMBER 2018
High mobility group 1 HMG1
 Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
RHOA inactivation enhances Wnt signaling and promotes colorectal cancer
 RHOA and colorectal cancer Paulo Rodrigues et al 2014 SUPPLEMENTARY INFORMATION RHOA inactivation enhances Wnt signaling and promotes colorectal cancer Paulo Rodrigues, Irati Macaya, Sarah Bazzocco, Rocco
RHOA and colorectal cancer Paulo Rodrigues et al 2014 SUPPLEMENTARY INFORMATION RHOA inactivation enhances Wnt signaling and promotes colorectal cancer Paulo Rodrigues, Irati Macaya, Sarah Bazzocco, Rocco
Table S2. Sequences immunoprecipitating with anti-hif-1α
 Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Table S1A. Generic EMT signature for tumour
 Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
Supporting Information. Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka
 Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated
 Supplemental Figure legends Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated genes. a) Mitotic roles of Polo-like kinase. b) Cell cycle regulation by BTG
Supplemental Figure legends Supplemental Figure 1. Pathways associated with the most significantly altered FGF-8b-regulated genes. a) Mitotic roles of Polo-like kinase. b) Cell cycle regulation by BTG
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells
 Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee
Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΒΙΟΧΗΜΕΙΑΣ & ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test Επιμέλεια
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΒΙΟΧΗΜΕΙΑΣ & ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test Επιμέλεια
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
Chapter 1. Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes
 Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Κυτταρική Επικοινωνία Είναι η ικανότητα του κυττάρου να αποκρίνεται σε εξωτερικά σήματα (σήματα από το περιβάλλον του ή άλλα κύτταρα) Τα σήματα αυτά μπορεί να είναι
Κυτταρική Επικοινωνία Cell Communication Κυτταρική Επικοινωνία Είναι η ικανότητα του κυττάρου να αποκρίνεται σε εξωτερικά σήματα (σήματα από το περιβάλλον του ή άλλα κύτταρα) Τα σήματα αυτά μπορεί να είναι
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor
 Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Differentially expressed genes of HEK293/HeLa Myst4 sirna vs. controls.
 Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance
 POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
5- CACGAAACTACCTTCAACTCC-3 beta actin-r 5- CATACTCCTGCTTGCTGATC-3 GAPDH-F GAPDH-R
 Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
A rearrangement is established as a barrier in parapatry. The accumulation of incompatibilites starts. Chrom 1. Rearranged
 Figure S1. An ancestral species splits in two according to the model discussed in Navarro and Barton (Evolution. In press. 2003). A rearrangement in chromosome 1 was involved in the process. From the moment
Figure S1. An ancestral species splits in two according to the model discussed in Navarro and Barton (Evolution. In press. 2003). A rearrangement in chromosome 1 was involved in the process. From the moment
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
 Supplemental Figure 1 Genechip data analysis. Three pairs of bulge and sub-bulge ORS Genechips were analyzed and compared by MASv5.0 and dchip 1.3 software algorithms in order to identify gene transcripts
Supplemental Figure 1 Genechip data analysis. Three pairs of bulge and sub-bulge ORS Genechips were analyzed and compared by MASv5.0 and dchip 1.3 software algorithms in order to identify gene transcripts
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Gene Primer sequence (5 to 3 ) Size (bp) E (%) GenBank
 Table S1 Primers used for quantitative real time PCR Gene Primer sequence (5 to 3 ) Size (bp) E (%) GenBank accession no: scd a got2a a ube2h a gpx4b a mknk2b a hsd11b2 b eef1a c b2m d F: ACCCGGAAGTCATCGAGAGA
Table S1 Primers used for quantitative real time PCR Gene Primer sequence (5 to 3 ) Size (bp) E (%) GenBank accession no: scd a got2a a ube2h a gpx4b a mknk2b a hsd11b2 b eef1a c b2m d F: ACCCGGAAGTCATCGAGAGA
Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver
 Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
 ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΦΥΣΙΟΛΟΓΙΑΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΕΡΓΑΣΤΗΡΙΟ ΠΕΙΡΑΜΑΤΙΚΗΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΙΕΥΘΥΝΤΡΙΑ: ΚΑΘΗΓΗΤΡΙΑ ΒΑΣΙΛΙΚΗ ΜΗΡΤΣΟΥ-ΦΙ ΑΝΗ ΠΑΝΕΠ. ΕΤΟΣ 2005-2006 ΑΡΙΘΜ. 1844 ΙΕΡΕΥΝΗΣΗ ΤΟΥ ΡΟΛΟΥ ΑΜΙΝΟΞΕΩΝ, ΠΕΠΤΙ
ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΦΥΣΙΟΛΟΓΙΑΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΕΡΓΑΣΤΗΡΙΟ ΠΕΙΡΑΜΑΤΙΚΗΣ ΦΑΡΜΑΚΟΛΟΓΙΑΣ ΙΕΥΘΥΝΤΡΙΑ: ΚΑΘΗΓΗΤΡΙΑ ΒΑΣΙΛΙΚΗ ΜΗΡΤΣΟΥ-ΦΙ ΑΝΗ ΠΑΝΕΠ. ΕΤΟΣ 2005-2006 ΑΡΙΘΜ. 1844 ΙΕΡΕΥΝΗΣΗ ΤΟΥ ΡΟΛΟΥ ΑΜΙΝΟΞΕΩΝ, ΠΕΠΤΙ
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL 1. Methods MicroRNA TaqMan Low Density Array MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Life Technologies). Global
SUPPLEMENTAL MATERIAL 1. Methods MicroRNA TaqMan Low Density Array MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Life Technologies). Global
Growth Factors/Wachstumsfaktoren. Human (Hu) Cytokines and Growth Factors/Humane (Hu) Cytokine und Wachstumsfaktoren
 Growth Factors/Wachstumsfaktoren Human (Hu) Cytokines and Growth Factors/Humane (Hu) Cytokine und Wachstumsfaktoren 4-1BBr Hu 4-1BB Receptor rec. CB-1010100 5 μg CB-1010101 20 μg CB-1010102 1 mg Acrp30
Growth Factors/Wachstumsfaktoren Human (Hu) Cytokines and Growth Factors/Humane (Hu) Cytokine und Wachstumsfaktoren 4-1BBr Hu 4-1BB Receptor rec. CB-1010100 5 μg CB-1010101 20 μg CB-1010102 1 mg Acrp30
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Διάλεξη 5. Μπράλιου Γεωργία, PhD, Τμήμα Πληροφορικής με Εφαρμογές στη Βιοϊατρική Πανεπιστήμιο Θεσσαλίας
 Διάλεξη 5 Δομή μεμβρανών (πάχος 5nm, 50 άτομα) Μεμβρανική μεταφορά Φραγμοί Μεμβρανικές πρωτεΐνες Λειτουργίες μεμβρανικών πρωτεϊνών Δίαυλοι-μεταφορείς-δυναμικό της μεμβράνης G protein coupled receptors
Διάλεξη 5 Δομή μεμβρανών (πάχος 5nm, 50 άτομα) Μεμβρανική μεταφορά Φραγμοί Μεμβρανικές πρωτεΐνες Λειτουργίες μεμβρανικών πρωτεϊνών Δίαυλοι-μεταφορείς-δυναμικό της μεμβράνης G protein coupled receptors
Supplementary Figure 1
 NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver.
 Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Supplemental tables and figures
 Supplemental tables and figures Supplemental Table S1. Demographic and clinical parameters by quartiles of white Variables blood cell count. WBC quartiles( 1 9 cells/l) Q1(.6-5.3, n=2587) Q2(5.4-6.3, n=263)
Supplemental tables and figures Supplemental Table S1. Demographic and clinical parameters by quartiles of white Variables blood cell count. WBC quartiles( 1 9 cells/l) Q1(.6-5.3, n=2587) Q2(5.4-6.3, n=263)
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
SUPPLEMENTARY INFORMATION. Pharmacological correction of a defect in PPAR-γ signaling ameliorates disease severity in Cftr-deficient mice
 SUPPLEMENTARY INFORMATION Pharmacological correction of a defect in PPAR-γ signaling ameliorates disease severity in Cftr-deficient mice Gregory S Harmon 1, Darren S Dumlao 2,3, Damian T Ng 4, Kim E Barrett
SUPPLEMENTARY INFORMATION Pharmacological correction of a defect in PPAR-γ signaling ameliorates disease severity in Cftr-deficient mice Gregory S Harmon 1, Darren S Dumlao 2,3, Damian T Ng 4, Kim E Barrett
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
Supplementary Information Titles
 Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
CRC Croce Tili List of patients for mir125b Metabolomic study Clinical data update 13/01/12
 AGGRESSIVE CRC Croce Tili List of patients for mir125b Metabolomic study Clinical data update 13/01/12 INDOLENT Table S1. Clinical data of the patients used for the study ID ZAP %VH WBC Dx date First TX
AGGRESSIVE CRC Croce Tili List of patients for mir125b Metabolomic study Clinical data update 13/01/12 INDOLENT Table S1. Clinical data of the patients used for the study ID ZAP %VH WBC Dx date First TX
Asrani et al Supplementary Figure S1: Epidermal-specific mtorc1 loss-of-function models show impaired epidermal differentiation.
 Asrani et al Supplementary Figure S1: Epidermal-specific mtorc1 loss-of-function models show impaired epidermal differentiation. (A) Rheb cko P0 epidermis fails to express spinous (Keratin 1, 10) and granular
Asrani et al Supplementary Figure S1: Epidermal-specific mtorc1 loss-of-function models show impaired epidermal differentiation. (A) Rheb cko P0 epidermis fails to express spinous (Keratin 1, 10) and granular
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine
 Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Table S1: Inpatient Diet Composition
 Table S1: Inpatient Diet Composition Diet Components (% of Energy) Inpatient Period Baseline Intervention Protein 15.1 ± 0.0 15.0 ± 0.1 Total fat 30.0 ± 0.0 30.0 ± 0.1 Saturated fat 8.6 ± 0.8 8.4 ± 0.9
Table S1: Inpatient Diet Composition Diet Components (% of Energy) Inpatient Period Baseline Intervention Protein 15.1 ± 0.0 15.0 ± 0.1 Total fat 30.0 ± 0.0 30.0 ± 0.1 Saturated fat 8.6 ± 0.8 8.4 ± 0.9
ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ
 ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΚΑΛΛΙΟΠΗ ΚΩΤΣΑ ΕΠΙΚ. ΚΑΘΗΓΗΤΡΙΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ Α.Π.Θ ΤΜΗΜΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ-ΔΙΑΒΗΤΟΛΟΓΙΚΟ
ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΚΑΛΛΙΟΠΗ ΚΩΤΣΑ ΕΠΙΚ. ΚΑΘΗΓΗΤΡΙΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ Α.Π.Θ ΤΜΗΜΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ-ΔΙΑΒΗΤΟΛΟΓΙΚΟ
Sar1B-mediated Regulation of Triglyceride and Cholesterol
 FIGURE S1. Comparing Sar1A- and Sar1B-flag Over-expression on RNA Profiles of McArdle RH7777 cells. RNA was prepared from two Sar1B:H79G-flag cell lines and two control cell lines plus two Sar1B-flag and
FIGURE S1. Comparing Sar1A- and Sar1B-flag Over-expression on RNA Profiles of McArdle RH7777 cells. RNA was prepared from two Sar1B:H79G-flag cell lines and two control cell lines plus two Sar1B-flag and
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
ΣΤΡΩΜΑΤΙΚΟΙ ΟΓΚΟΙ ΓΑΣΤΡΕΝΤΕΡΙΚΟΥ ΣΥΣΤΗΜΑΤΟΣ (GISTs) ΕΛΕΝΗ ΠΑΠΑΔΑΚΗ ΠΑΘΟΛΟΓΟΑΝΑΤΟΜΟΣ ΝΟΣΟΚΟΜΕΙΟ ΠΑΤΡΩΝ «ΑΓΙΟΣ ΑΝΔΡΕΑΣ»
 ΣΤΡΩΜΑΤΙΚΟΙ ΟΓΚΟΙ ΓΑΣΤΡΕΝΤΕΡΙΚΟΥ ΣΥΣΤΗΜΑΤΟΣ (GISTs) ΕΛΕΝΗ ΠΑΠΑΔΑΚΗ ΠΑΘΟΛΟΓΟΑΝΑΤΟΜΟΣ ΝΟΣΟΚΟΜΕΙΟ ΠΑΤΡΩΝ «ΑΓΙΟΣ ΑΝΔΡΕΑΣ» ΠΟΙΟΥΣ ΟΓΚΟΥΣ ΟΝΟΜΑΖΟΥΜΕ GIST? Τους μεσεγχυματικούς όγκους του ΓΕΣ,του περιτοναιου
ΣΤΡΩΜΑΤΙΚΟΙ ΟΓΚΟΙ ΓΑΣΤΡΕΝΤΕΡΙΚΟΥ ΣΥΣΤΗΜΑΤΟΣ (GISTs) ΕΛΕΝΗ ΠΑΠΑΔΑΚΗ ΠΑΘΟΛΟΓΟΑΝΑΤΟΜΟΣ ΝΟΣΟΚΟΜΕΙΟ ΠΑΤΡΩΝ «ΑΓΙΟΣ ΑΝΔΡΕΑΣ» ΠΟΙΟΥΣ ΟΓΚΟΥΣ ΟΝΟΜΑΖΟΥΜΕ GIST? Τους μεσεγχυματικούς όγκους του ΓΕΣ,του περιτοναιου
Supporting Information
 Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were
Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were
Η Α ΕΝΟΫΠΟΦΥΣΗ: OI ΓΟΝΑ ΟΤΡΟΠΙΝΕΣ (FSH, LH) ΚΑΙ Η ΠΡΟΛΑΚΤΙΝΗ (PRL)
 Η Α ΕΝΟΫΠΟΦΥΣΗ: OI ΓΟΝΑ ΟΤΡΟΠΙΝΕΣ (FSH, LH) ΚΑΙ Η ΠΡΟΛΑΚΤΙΝΗ (PRL) Κωνσταντίνος Καλλαράς Ιατρός Παθολόγος Καθηγητής Φυσιολογίας Εργαστήριο Πειραματικής Φυσιολογίας Ιατρικής Σχολής Α.Π.Θ. FSH:ΜΒ 33000,
Η Α ΕΝΟΫΠΟΦΥΣΗ: OI ΓΟΝΑ ΟΤΡΟΠΙΝΕΣ (FSH, LH) ΚΑΙ Η ΠΡΟΛΑΚΤΙΝΗ (PRL) Κωνσταντίνος Καλλαράς Ιατρός Παθολόγος Καθηγητής Φυσιολογίας Εργαστήριο Πειραματικής Φυσιολογίας Ιατρικής Σχολής Α.Π.Θ. FSH:ΜΒ 33000,
Figure S1. Localization of GiOR-1 with truncated N-terminal domain of 29 amino acid residues (ΔL-GiOR-1-HA).
 Figure S1. Localization of GiOR-1 with truncated N-terminal domain of 29 amino acid residues (ΔL-GiOR-1-HA). A L-GiOR-1-HA Tom40 DAPI merge DIC B LYS CYT ORG ORG Trypsin - - - + L-GiOR-1-HA Figure S2A.
Figure S1. Localization of GiOR-1 with truncated N-terminal domain of 29 amino acid residues (ΔL-GiOR-1-HA). A L-GiOR-1-HA Tom40 DAPI merge DIC B LYS CYT ORG ORG Trypsin - - - + L-GiOR-1-HA Figure S2A.
Guo_Fig. S1. Atg7 +/+ Atg7 -/- FBP_M+6 DHAP_M+3. Glucose Uptake Rate G6P_M+6. nmol/ul cell/hr
 Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Transcriptional signaling pathways inversely regulated in Alzheimer s disease and glioblastoma multiform
 Transcriptional signaling pathways inversely regulated in Alzheimer s disease and glioblastoma multiform Timothy Liu, Ding Ren, Xiaoping Zhu, Zheng Yin, Guangxu Jin, Zhen Zhao, Daniel Robinson, Xuping
Transcriptional signaling pathways inversely regulated in Alzheimer s disease and glioblastoma multiform Timothy Liu, Ding Ren, Xiaoping Zhu, Zheng Yin, Guangxu Jin, Zhen Zhao, Daniel Robinson, Xuping
Μελέτη της επίδρασης αντικαρκινικών χημειοθεραπευτικών φαρμάκων στη ρύθμιση του Cdt1
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραμμα Μεταπτυχιακών Σπουδών Εφαρμογές Στις Βασικές Επιστήμες Κατεύθυνση: Φαρμακοκινητική Τοξικολογία Διπλωματική Εργασία Μελέτη της επίδρασης
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραμμα Μεταπτυχιακών Σπουδών Εφαρμογές Στις Βασικές Επιστήμες Κατεύθυνση: Φαρμακοκινητική Τοξικολογία Διπλωματική Εργασία Μελέτη της επίδρασης
Growth Factors/Wachstumsfaktoren. Human (Hu) Cytokines and Growth Factors/Humane (Hu) Cytokine und Wachstumsfaktoren
 4-1BBr Hu 4-1BB Receptor rec. CB-1010100 5 μg CB-1010101 20 μg CB-1010102 1 mg Acrp30 Glob Hu Adiponectin Globular rec. CB-2800004 2 μg CB-2800005 10 μg CB-280000 1 mg Acrp30 HEK Hu Adiponectin glycosilated,
4-1BBr Hu 4-1BB Receptor rec. CB-1010100 5 μg CB-1010101 20 μg CB-1010102 1 mg Acrp30 Glob Hu Adiponectin Globular rec. CB-2800004 2 μg CB-2800005 10 μg CB-280000 1 mg Acrp30 HEK Hu Adiponectin glycosilated,
Επασβεστώσεις βασικών γαγγλίων και status epilepticus σε παιδί με ψευδοϋποπαραθυρεοειδισμό τύπου Ia
 Παιδιατρική ΒΟΡΕΙΟΥ Ε ΛΛΑ ΔΟΣ, 25: 65-71, 2013 Παιδιατρική ΒΟΡΕΙΟΥ ΕΛΛΑΔΟΣ, 25 1 65 Επασβεστώσεις βασικών γαγγλίων και status epilepticus σε παιδί με ψευδοϋποπαραθυρεοειδισμό τύπου Ia Β. Κούγια 1, Φ. Αδαμίδου
Παιδιατρική ΒΟΡΕΙΟΥ Ε ΛΛΑ ΔΟΣ, 25: 65-71, 2013 Παιδιατρική ΒΟΡΕΙΟΥ ΕΛΛΑΔΟΣ, 25 1 65 Επασβεστώσεις βασικών γαγγλίων και status epilepticus σε παιδί με ψευδοϋποπαραθυρεοειδισμό τύπου Ia Β. Κούγια 1, Φ. Αδαμίδου
15 A-NET ( ) 1 3 Mesoderm 2 4 Ectoderm 3 5 Mesoderm 4 6 endoderm. 1. Fe, s 2. Mg, N 3. Fe, N 4. Mg, S
 122 15 A-NET ( ) 1. ก ก blood vessel ก 1 Mesoderm 2 4 Ectoderm 5 Mesoderm 4 6 endoderm 2. กก ก. ก. ก ก. ก. ก ก ก 1. ก 2.. ก, 4.,. ก 1. Fe, s 2. Mg, N. Fe, N 4. Mg, S 4. ก กก ก ก ก ก ก ก 1. ก ก ก ก กก 2.
122 15 A-NET ( ) 1. ก ก blood vessel ก 1 Mesoderm 2 4 Ectoderm 5 Mesoderm 4 6 endoderm 2. กก ก. ก. ก ก. ก. ก ก ก 1. ก 2.. ก, 4.,. ก 1. Fe, s 2. Mg, N. Fe, N 4. Mg, S 4. ก กก ก ก ก ก ก ก 1. ก ก ก ก กก 2.
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Chalkou I. C. [PROJECT] Ανάθεση εργασιών.
![Chalkou I. C. [PROJECT] Ανάθεση εργασιών. Chalkou I. C. [PROJECT] Ανάθεση εργασιών.](/thumbs/29/13219375.jpg) Πληροφορική της Υγείας 2014 Chalkou I. C. [PROJECT] Ανάθεση εργασιών. Περιεχόμενα 1. Ομάδα ΣΤ... 3 1.1 ΜΑΡΚΟΠΟΥΛΟΥ- ΣΠΥΡΟΠΟΥΛΟΥ -ΚΩΝΣΤΑΝΤΟΠΟΥΛΟΥ... 3 1.2 ΜΑΡΚΟΣ- ΚΟΥΤΣΟΠΟΥΛΟΣ ΑΥΓΕΡΗ - ΜΠΟΥΖΑΛΑ... 3 1.3
Πληροφορική της Υγείας 2014 Chalkou I. C. [PROJECT] Ανάθεση εργασιών. Περιεχόμενα 1. Ομάδα ΣΤ... 3 1.1 ΜΑΡΚΟΠΟΥΛΟΥ- ΣΠΥΡΟΠΟΥΛΟΥ -ΚΩΝΣΤΑΝΤΟΠΟΥΛΟΥ... 3 1.2 ΜΑΡΚΟΣ- ΚΟΥΤΣΟΠΟΥΛΟΣ ΑΥΓΕΡΗ - ΜΠΟΥΖΑΛΑ... 3 1.3
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
JCVI Role Category_1 JCVI Role Category_2. histidinol phosphatase-related protein Unknown function NT01ST0303 AAL19211 STM0248
 Additional Table 3. Detailed information on all 81 genes that showed evidence for positive selection from initial positive selection analysis, in Protein Name Gene name JCVI Role Category_1 JCVI Role Category_2
Additional Table 3. Detailed information on all 81 genes that showed evidence for positive selection from initial positive selection analysis, in Protein Name Gene name JCVI Role Category_1 JCVI Role Category_2
Το Βιολογικό Ρολόι: Θεµελιώδης Ρυθµιστής της Φυσιολογίας των Οργανισµών
 Ιούλιος 2005 Το Βιολογικό Ρολόι: Θεµελιώδης Ρυθµιστής της Φυσιολογίας των Οργανισµών ρ. Α. Προµπονά Εργαστήριο Ρύθµισης της Μεταγραφής από το Βιολογικό Ρολόι Κιρκαδικό Ρολόι Ρυθµοί µε περίοδο ~ 24 ωρών
Ιούλιος 2005 Το Βιολογικό Ρολόι: Θεµελιώδης Ρυθµιστής της Φυσιολογίας των Οργανισµών ρ. Α. Προµπονά Εργαστήριο Ρύθµισης της Μεταγραφής από το Βιολογικό Ρολόι Κιρκαδικό Ρολόι Ρυθµοί µε περίοδο ~ 24 ωρών
JCVI Locus Protein Name. JCVI Role Category JCVI Locus (LT2) Annotation name
 Additional File 4. Detailed information on all 41 genes that showed evidence for positive selection from positive selection analysis, in which gene Alignment Primary Gene Genbank Locus JCVI Locus Protein
Additional File 4. Detailed information on all 41 genes that showed evidence for positive selection from positive selection analysis, in which gene Alignment Primary Gene Genbank Locus JCVI Locus Protein
Ινσουλίνη και γήρανση
 Ινσουλίνη και γήρανση Γεώργιος Δ. Δημητριάδης Καθηγητής Παθολογίας Διευθυντής Β Προπαιδευτικής Παθολογικής Κλινικής, Μονάδας Έρευνας και Διαβητολογικού Κέντρου Πανεπιστημίου Αθηνών, ΠΓΝΑ Αττικόν Πρόεδρος
Ινσουλίνη και γήρανση Γεώργιος Δ. Δημητριάδης Καθηγητής Παθολογίας Διευθυντής Β Προπαιδευτικής Παθολογικής Κλινικής, Μονάδας Έρευνας και Διαβητολογικού Κέντρου Πανεπιστημίου Αθηνών, ΠΓΝΑ Αττικόν Πρόεδρος
Αντιδράσεις οξειδωτικού μέρους. Μη οξειδωτικές αντιδράσεις ΔΡΟΜΟΣ ΤΩΝ ΦΩΣΦΟΤΙΚΩΝ ΠΕΝΤΟΖΩΝ. 6-Ρ γλυκόζη. 5-Ρ ριβουλόζη
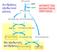 Αντιδράσεις οξειδωτικού μέρους 6-Ρ γλυκόζη 5-Ρ ριβουλόζη ΔΡΟΜΟΣ ΤΩΝ ΦΩΣΦΟΤΙΚΩΝ ΠΕΝΤΟΖΩΝ 5-Ρ ριβόζη (C 5 ) 5-Ρ ξυλουλόζη (C 5 ) GAP (C 3 ) 7-Ρ σεδοεπτουλόζη (C 7 ) 6-Ρ φρουκτόζη(c 6 ) 4-Ρ ερυθρόζη (C 5
Αντιδράσεις οξειδωτικού μέρους 6-Ρ γλυκόζη 5-Ρ ριβουλόζη ΔΡΟΜΟΣ ΤΩΝ ΦΩΣΦΟΤΙΚΩΝ ΠΕΝΤΟΖΩΝ 5-Ρ ριβόζη (C 5 ) 5-Ρ ξυλουλόζη (C 5 ) GAP (C 3 ) 7-Ρ σεδοεπτουλόζη (C 7 ) 6-Ρ φρουκτόζη(c 6 ) 4-Ρ ερυθρόζη (C 5
Supplementary Table 1. Demographic characteristics of the included ADNI subjects. rfmri. MMSE tics (n=907; (n=1092; n long =107.
 Supplementary Fig. 1. Assumed disease state (DS) vs aging relationships. In a) the DS increases linearly with age, while in b) it follows a sigmoid relationship with age (as suggested by 1 ). In both cases,
Supplementary Fig. 1. Assumed disease state (DS) vs aging relationships. In a) the DS increases linearly with age, while in b) it follows a sigmoid relationship with age (as suggested by 1 ). In both cases,
BD Cytometric Bead Array Flex Set System
 BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
The NF-κB pathways in connective tissue diseases
 κ The NF-κB pathways in connective tissue diseases TETSUO KUBOTA (Associate Professor) Tokyo Medical and Dental University Graduate School of Health Care Sciences NF-κB nuclear factor-kappa B NF-κB in
κ The NF-κB pathways in connective tissue diseases TETSUO KUBOTA (Associate Professor) Tokyo Medical and Dental University Graduate School of Health Care Sciences NF-κB nuclear factor-kappa B NF-κB in
ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ
 5 ο ΠΑΝΕΛΛΗΝΙΟ ΣΥΝΕΔΡΙΟ ΕΛΛΗΝΙΚΗΣ ΕΤΑΙΡΕΙΑΣ ΑΙΜΑΦΑΙΡΕΣΗΣ ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ Θωμάς Τσαγανός, MD, PhD Πανεπιστημιακός Υπότροφος Δ Παθολογική Κλινική Πανεπιστημιακό Γενικό Νοσοκομείο
5 ο ΠΑΝΕΛΛΗΝΙΟ ΣΥΝΕΔΡΙΟ ΕΛΛΗΝΙΚΗΣ ΕΤΑΙΡΕΙΑΣ ΑΙΜΑΦΑΙΡΕΣΗΣ ΓΕΝΙΚΕΣ ΑΡΧΕΣ ΑΝΟΣΟΠΑΘΟΓΕΝΕΙΑΣ ΣΤΗ ΣΗΨΗ Θωμάς Τσαγανός, MD, PhD Πανεπιστημιακός Υπότροφος Δ Παθολογική Κλινική Πανεπιστημιακό Γενικό Νοσοκομείο
ΠΑΤΡΩΝΥΜΟ / ΟΝΟΜΑ ΣΥΖΥΓΟΥ 1 ΑΓΟΡΑΣΤΟΥ ΜΑΡΙΑ ΤΟΥ ΔΗΜΗΤΡΙΟΥ 2 ΑΘΑΝΑΣΙΑΔΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΠΑΥΛΟΥ 3 ΑΚΤΣΟΓΛΟΥ ΣΩΚΡΑΤΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ
 Υποψήφιοι ημοτικοί Σύμβουλοι: ΠΑΤΡΩΝΥΜΟ / ΣΥΖΥΓΟΥ 1 ΑΓΟΡΑΣΤΟΥ ΜΑΡΙΑ ΤΟΥ ΔΗΜΗΤΡΙΟΥ 2 ΑΘΑΝΑΣΙΑΔΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΠΑΥΛΟΥ 3 ΑΚΤΣΟΓΛΟΥ ΣΩΚΡΑΤΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 4 ΑΛΦΑΤΖΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 5 ΑΜΟΡΓΙΑΝΟΣ ΝΙΚΟΛΑΟΣ
Υποψήφιοι ημοτικοί Σύμβουλοι: ΠΑΤΡΩΝΥΜΟ / ΣΥΖΥΓΟΥ 1 ΑΓΟΡΑΣΤΟΥ ΜΑΡΙΑ ΤΟΥ ΔΗΜΗΤΡΙΟΥ 2 ΑΘΑΝΑΣΙΑΔΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΠΑΥΛΟΥ 3 ΑΚΤΣΟΓΛΟΥ ΣΩΚΡΑΤΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 4 ΑΛΦΑΤΖΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 5 ΑΜΟΡΓΙΑΝΟΣ ΝΙΚΟΛΑΟΣ
Supporting Information
 Supporting Information Aluminum Complexes of N 2 O 2 3 Formazanate Ligands Supported by Phosphine Oxide Donors Ryan R. Maar, Amir Rabiee Kenaree, Ruizhong Zhang, Yichen Tao, Benjamin D. Katzman, Viktor
Supporting Information Aluminum Complexes of N 2 O 2 3 Formazanate Ligands Supported by Phosphine Oxide Donors Ryan R. Maar, Amir Rabiee Kenaree, Ruizhong Zhang, Yichen Tao, Benjamin D. Katzman, Viktor
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Τι το νεότερο στη φαρμακευτική θεραπεία; Παραμένουν οι στατίνες ως φάρμακα επιλογής;
 Τι το νεότερο στη φαρμακευτική θεραπεία; Παραμένουν οι στατίνες ως φάρμακα επιλογής; Άννα Ταυρίδου,, PhD Εργαστήριο Φαρμακολογίας, Τμήμα Ιατρικής, ημοκρίτειο Πανεπιστήμιο Θράκης Υπάρχει ανάγκη για καινούργια
Τι το νεότερο στη φαρμακευτική θεραπεία; Παραμένουν οι στατίνες ως φάρμακα επιλογής; Άννα Ταυρίδου,, PhD Εργαστήριο Φαρμακολογίας, Τμήμα Ιατρικής, ημοκρίτειο Πανεπιστήμιο Θράκης Υπάρχει ανάγκη για καινούργια
Supporting Information
 Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2013 Structure Activity Relationship Study Reveals ML240 and ML241 as Potent and Selective Inhibitors of p97 ATPase Tsui-Fen
Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2013 Structure Activity Relationship Study Reveals ML240 and ML241 as Potent and Selective Inhibitors of p97 ATPase Tsui-Fen
ΠΕΡΙΛΗΨΗ ΧΑΡΑΚΤΗΡΙΣΤΙΚΩΝ ΤΟΥ ΠΡΟΙΟΝΤΟΣ (SPC) (AMINOVEN 3.5% Glucose / Electrolytes)
 ΠΕΡΙΛΗΨΗ ΧΑΡΑΚΤΗΡΙΣΤΙΚΩΝ ΤΟΥ ΠΡΟΙΟΝΤΟΣ (SPC) (AMINOVEN 3.5% Glucose / Electrolytes) 1. ΕΜΠΟΡΙΚΗ ΟΝΟΜΑΣΙΑ ΤΟΥ ΦΑΡΜΑΚΕΥΤΙΚΟΥ ΠΡΟΙΟΝΤΟΣ: AMINOVEN 3.5% Glucose / Electrolytes 2. ΠΟΙΟΤΙΚΗ ΚΑΙ ΠΟΣΟΤΙΚΗ ΣΥΝΘΕΣΗ
ΠΕΡΙΛΗΨΗ ΧΑΡΑΚΤΗΡΙΣΤΙΚΩΝ ΤΟΥ ΠΡΟΙΟΝΤΟΣ (SPC) (AMINOVEN 3.5% Glucose / Electrolytes) 1. ΕΜΠΟΡΙΚΗ ΟΝΟΜΑΣΙΑ ΤΟΥ ΦΑΡΜΑΚΕΥΤΙΚΟΥ ΠΡΟΙΟΝΤΟΣ: AMINOVEN 3.5% Glucose / Electrolytes 2. ΠΟΙΟΤΙΚΗ ΚΑΙ ΠΟΣΟΤΙΚΗ ΣΥΝΘΕΣΗ
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplementary Information
 Supplementary Information Supplementary Figure 1: Characterization of cell surface biotinylation in non-tumorigenic and tumorigenic HCC cell lines. (a) Indicated cell lines were either mock or surface
Supplementary Information Supplementary Figure 1: Characterization of cell surface biotinylation in non-tumorigenic and tumorigenic HCC cell lines. (a) Indicated cell lines were either mock or surface
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
BioVision Catalog Description Pack Size Supplier Price (RS)
 Catalog Description Pack Size Supplier Price (RS) K101-100 Annexin V-FITC Apoptosis Kit 100 assays BioVision 60991 K101-25 Annexin V-FITC Apoptosis Kit 25 assays BioVision 28373 K101-400 Annexin V-FITC
Catalog Description Pack Size Supplier Price (RS) K101-100 Annexin V-FITC Apoptosis Kit 100 assays BioVision 60991 K101-25 Annexin V-FITC Apoptosis Kit 25 assays BioVision 28373 K101-400 Annexin V-FITC
Lclat Agpat Lpcat Agpat Lpcat Lpcat Lpcat Lpgat Awat1 N.D. Mboat Awat2 N.D.
 Supplemental Table I. The gene expression of 7 acyltransferases in MOVAS-1 cells. Gene Name (pg)/18s(ng) Gene Name (pg)/18s(ng) Acat1 19.53 Gpat 9.75 Acat. Lclat1 1.5 Agpat1 3.31 Lpcat1 8.15 Agpat.16 Lpcat.
Supplemental Table I. The gene expression of 7 acyltransferases in MOVAS-1 cells. Gene Name (pg)/18s(ng) Gene Name (pg)/18s(ng) Acat1 19.53 Gpat 9.75 Acat. Lclat1 1.5 Agpat1 3.31 Lpcat1 8.15 Agpat.16 Lpcat.
Ο ρόλος της αιμαφαίρεσης στην μεταμόσχευση αιμοποιητικών κυττάρων. Παναγιώτης Τσιριγώτης
 Ο ρόλος της αιμαφαίρεσης στην μεταμόσχευση αιμοποιητικών κυττάρων Παναγιώτης Τσιριγώτης Αιμαφαίρεση και Μεταμόσχευση Λήψη του μοσχεύματος Αντιμετώπιση επιπλοκών Αντιμετώπιση νόσου του μοσχεύματος έναντι
Ο ρόλος της αιμαφαίρεσης στην μεταμόσχευση αιμοποιητικών κυττάρων Παναγιώτης Τσιριγώτης Αιμαφαίρεση και Μεταμόσχευση Λήψη του μοσχεύματος Αντιμετώπιση επιπλοκών Αντιμετώπιση νόσου του μοσχεύματος έναντι
Αθήνα, 15 Οκτωβρίου 2014 Αρ. Πρωτ.: 2988
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΥΠΟΥΡΓΕΙΟ ΠΑΙΔΕΙΑΣ ΚΑΙ ΘΡΗΣΚΕΥΜΑΤΩΝ ΓΕΝΙΚΗ ΓΡΑΜΜΑΤΕΙΑ ΕΡΕΥΝΑΣ & ΤΕΧΝΟΛΟΓΙΑΣ Αθήνα, 15 Οκτωβρίου 2014 Αρ. Πρωτ.: 2988 Θέμα: Έγκριση πρόσκλησης εκδήλωσης ενδιαφέροντος με απευθείας ανάθεση
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΥΠΟΥΡΓΕΙΟ ΠΑΙΔΕΙΑΣ ΚΑΙ ΘΡΗΣΚΕΥΜΑΤΩΝ ΓΕΝΙΚΗ ΓΡΑΜΜΑΤΕΙΑ ΕΡΕΥΝΑΣ & ΤΕΧΝΟΛΟΓΙΑΣ Αθήνα, 15 Οκτωβρίου 2014 Αρ. Πρωτ.: 2988 Θέμα: Έγκριση πρόσκλησης εκδήλωσης ενδιαφέροντος με απευθείας ανάθεση
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
19 Ενδοκρινολόγος. Φάρµακα που στοχεύουν το β- κύτταρο ΕΥΡΥΔΙΚΗ ΠΑΠΑΔΟΠΟΥΛΟΥ ΕΙΣΑΓΩΓΗ ΜΗΧΑΝΙΣΜΟΣ ΔΡΑΣΗΣ
 Φάρµακα που στοχεύουν το β- κύτταρο ΕΥΡΥΔΙΚΗ ΠΑΠΑΔΟΠΟΥΛΟΥ 19 Ενδοκρινολόγος ΕΙΣΑΓΩΓΗ Οι σουλφονυλουρίες είναι η πρώτη κατηγορία υπογλυκαιμικών φαρμάκων που χορηγήθηκαν για την αντιμετώπιση του διαβήτη
Φάρµακα που στοχεύουν το β- κύτταρο ΕΥΡΥΔΙΚΗ ΠΑΠΑΔΟΠΟΥΛΟΥ 19 Ενδοκρινολόγος ΕΙΣΑΓΩΓΗ Οι σουλφονυλουρίες είναι η πρώτη κατηγορία υπογλυκαιμικών φαρμάκων που χορηγήθηκαν για την αντιμετώπιση του διαβήτη
SUPPLEMENTARY INFORMATION
 Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
ΙΑΤΑΡΑΧΕΣ ΜΕΤΑΒΟΛΙΣΜΟΥ ΛΙΠΑΡΩΝ ΟΞΕΩΝ
 Ηµ/νία Εξέτασης : ΙΑΤΑΡΑΧΕΣ ΜΕΤΑΒΟΛΙΣΜΟΥ ΛΙΠΑΡΩΝ ΟΞΕΩΝ Medium Chain Acyl CoA Dehydrogenase C6< 0.24 C8< 0.35 C10< 0.3 C10:1 < 0.22 C8/C2< 0.02 C8/C10 < 3 Long Chain 3 Hydroxyacyl Coa Dehydrogenase C16OH
Ηµ/νία Εξέτασης : ΙΑΤΑΡΑΧΕΣ ΜΕΤΑΒΟΛΙΣΜΟΥ ΛΙΠΑΡΩΝ ΟΞΕΩΝ Medium Chain Acyl CoA Dehydrogenase C6< 0.24 C8< 0.35 C10< 0.3 C10:1 < 0.22 C8/C2< 0.02 C8/C10 < 3 Long Chain 3 Hydroxyacyl Coa Dehydrogenase C16OH
