DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial
|
|
|
- Άρχέλαος Μνάσων Κανακάρης-Ρούφος
- 6 χρόνια πριν
- Προβολές:
Transcript
1 DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial pathogenicity on rice Jianmei Su 1, Xia Zou 1, Liangbo Huang 1, Tenglong Bai 1, Shu Liu 1, Meng Yuan 2, Shan-Ho Chou 3, Ya-Wen He 4, Haihong Wang 5, Jin He 1 *. 1 State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei , China. 2 National Key Laboratory of Crop Genetic Improvement, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei , China. 3 Institute of Biochemistry, and NCHU Agricultural Biotechnology Center, National Chung Hsing University, Taichung 40227, Taiwan. 4 State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai , China. 5 College of Life Sciences, South China Agricultural University, Guangzhou, Guangdong , China. *Corresponding author: hejin@mail.hzau.edu.cn (JH) 1
2 Section A: supplementary figures Figure S1. Multidomain architectures of DGCs and sequence alignment of DgcA with XCC3486. (a) Architectures of GGDEF domain-containing proteins with a GGDEF or GGEEF motifs in the Xoo genome. Predicted TM regions are shown as orange rectangles. (b) The amino acid sequence alignment of DgcA (XOO3988) in this study with XCC3486 from Xcc. 2
3 Figure S2. Purified tdgca is active as a DGC in vitro. (a) SDS-PAGE analysis of purified tdgca. Lane M: molecular markers; Lane 1: purified tdgca. (b), (c) and (d) were the HPLC chromatograms of the in vitro reaction products of tdgca, the GTP standard and c-di-gmp standard, respectively. 3
4 Figure S3. Construction and verification of the WT, ΔdgcA, OdgcA and CdgcA strains. (a) Schematic diagram of the gene deletion construct. Allelic homologous recombination strategy was applied with the suicide vector pk18mobsacb by replacing dgca with Gm gene. (b) PCR analyses of the ΔdgcA and CdgcA strains. Products were amplified using primer pair Gm-F and dgca-down-r from ΔdgcA (lanes 1-2) and CdgcA strains (lanes 3-4) because both strains contained a replaced Gm gene; M: DL2000 DNA ladder. (c) PCR confirmation of the WT, ΔdgcA, OdgcA and CdgcA strains. Products were amplified using primer pair Gm-F and Gm-R from WT (lanes 1-2), ΔdgcA (lanes 3-4), CdgcA (lanes 5-6) and OdgcA (lanes 7-8) strains, respectively. Meanwhile, since OdgcA and CdgcA strains harbored a kan gene from pbbr1mcs-dgca plasmid, another primer pair Kan-F and Kan-R was also used to amplify products from WT (lanes 9-10), ΔdgcA (lanes 11-12), CdgcA (lanes 13-14) and OdgcA (lanes 15-16) strains, respectively. M: DL2000 DNA ladder. (d) Verification of the DgcA protein in the CdgcA and OdgcA strains by Western blotting assay with the anti-his mouse monoclonal antibody. M: protein marker; Lane 1: supernatant of CdgcA; Line 2: supernatant of OdgcA; Line 3: supernatant sample of WT. 4
5 Figure S4. In Planta virulence test of the WT, ΔdgcA, OdgcA, CdgcA and Xoo-pBBR strains. (a) Leaf segments of rice cultivar Zhonghua 11 (ZH11) showing rice blight symptoms. Typical leaves were photographed after infection for 12 days with the four different strains. (b) Measurement of the mean lesion lengths 12 days post inoculation. Leaf without bacterial inoculation were served as a control. (c) Determination of bacterial population in rice leaves at four specific time points (3, 6, 9, 12 days after inoculation). The values were means ± standard deviations for triplicate assays. Significances of differences by Student s t-test are indicated (**P<0.01; *P<0.05; NS, P>0.05). 5
6 Figure S5. GO tree displays enrich genes associated with certain biological process. (a) GO enrichment of up-regulated DGEs. (b) GO enrichment of down-regulated DGEs. The color of the nodes indicates their significance according to the p value. 6
7 Figure S6. Mapping information of dgca (XOO3988) and its downstream gene pepn (XOO3989). The clean reads of WT and ΔdgcA were well mapped to the genome by Artemis software. 7
8 Section B: supplementary tables Sample Raw_reads Before clean After clean Kept_reads WT Xoo-wt-F.fq % Xoo-wt-R.fq % dgca Xoo dgca-f.fq % Xoo dgca-r.fq % Table S1. Statistics of reads that cleaned up. 8
9 gene locus_tag function length ΔdgcA WT ΔdgcA/WT ΔdgcA/WT GeneID protein_id product gi reads RPKM reads RPKM log2 (Fold_ change) normalized q-value result gi ref YP_ XOO0006 hypothetical protein E-16 down YP_ gi ref YP_ XOO0029 TonB-dependent receptor E-17 down YP_ iron gi ref YP_ XOO0032 transposase E-149 up YP_ gi ref YP_ XOO0111 hypothetical protein E-60 down YP_ gi ref YP_ XOO0119 methylamine utilization protein E-22 up YP_ maug gi ref YP_ XOO0122 transposase E-168 up YP_ gi ref YP_ XOO0131 VirK protein down YP_ virk gi ref YP_ XOO0136 transposase E-54 up YP_ gi ref YP_ XOO0161 transposase E-138 up YP_ gi ref YP_ XOO0166 ATP-dependent RNA helicase E-61 up YP_ gi ref YP_ XOO0167 hypothetical protein E-12 up YP_ gi ref YP_ XOO0168 avirulence protein E-33 down YP_ avrbs gi ref YP_ XOO0181 IS30 family transposase E-08 down YP_ gi ref YP_ XOO0182 transposase E-208 up YP_ gi ref YP_ XOO0200 transposase E-68 up YP_ gi ref YP_ XOO0201 transposase E-27 up YP_ gi ref YP_ XOO0282 cellulase E-144 down YP_ egl gi ref YP_ XOO0283 cellulase E-250 down YP_ egl gi ref YP_ XOO0297 transposase E-47 up YP_ gi ref YP_ XOO0300 transposase E-251 up YP_ gi ref YP_ XOO0312 acetyltransferase E-20 up YP_ gi ref YP_ XOO0313 transposase E-219 up YP_ gi ref YP_ XOO0342 transposase E-142 up YP_ gi ref YP_ XOO0414 transposase E-111 up YP_ gi ref YP_ XOO0415 transposase up YP_ gi ref YP_ XOO0416 membrane-fusion protein E-20 up YP_ gi ref YP_ XOO0417 catalase down YP_ catb gi ref YP_ XOO0418 ankyrin-like protein E-21 down YP_ ankb gi ref YP_ XOO0419 hypothetical protein E-98 down YP_ gi ref YP_ XOO0545 transposase up YP_ gi ref YP_ XOO0546 transposase E-101 up YP_ gi ref YP_ XOO0561 C-type cytochrome biogenesis protein (copper tolerance) E-28 up YP_ dsbd gi ref YP_ XOO0563 transposase E-187 up YP_ gi ref YP_ XOO0611 hypothetical protein E-44 up YP_
10 gi ref YP_ XOO0620 transposase E-165 up YP_ gi ref YP_ XOO0669 hypothetical protein E-51 up YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO0670 XOO0687 XOO0688 DNA polymerase related protein E-38 up YP_ flagellar motor protein MotB E-84 down YP_ motb flagellar motor protein MotA E-67 down YP_ mota gi ref YP_ XOO0690 methionine sulfoxide reductase B up YP_ gi ref YP_ XOO0716 hypothetical protein up YP_ gi ref YP_ XOO0737 histidine kinaseresponse regulator hybrid protein up YP_ stys gi ref YP_ XOO0739 transposase E-138 up YP_ gi ref YP_ XOO0747 hypothetical protein E-88 down YP_ gi ref YP_ XOO0748 hypothetical protein down YP_ gi ref YP_ XOO0786 IS1478 transposase down YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO0793 XOO0794 XOO0795 glucose-1-phosphate thymidylyltransferas e E-27 up YP_ rmla dtdp-4- dehydrorhamnose 3,5-epimerase E-31 up YP_ rmlc dtdp-4- dehydrorhamnose reductase E-26 up YP_ rmld gi ref YP_ XOO0812 gi ref YP_ XOO0853 ribosomal-proteinalanine acetyltransferase down YP_ rimi general secretion pathway protein K E-16 down YP_ xpsk gi ref YP_ XOO0874 transposase E-118 up YP_ gi ref YP_ XOO0875 transposase up YP_ gi ref YP_ XOO0879 hypothetical protein down YP_ gi ref YP_ XOO0917 IS1595 transposase E-104 up YP_ gi ref YP_ XOO0932 transposase E-95 up YP_ gi ref YP_ XOO0956 transposase E-206 up YP_ gi ref YP_ XOO0957 H+ translocating pyrophosphate synthase E-77 down YP_ gi ref YP_ XOO0960 transposase E-152 up YP_ gi ref YP_ XOO0979 disulfide bond formation protein B E-11 down YP_ dsbb gi ref YP_ XOO0980 IS1478 transposase up YP_ gi ref YP_ XOO0989 transposase E-87 up YP_ gi ref YP_ XOO1003 transposase E-135 up YP_ gi ref YP_ XOO1023 transposase E-185 up YP_ gi ref YP_ XOO1053 transposase E-141 up YP_ gi ref YP_ XOO1062 molybdopterinconverting factor subunit down YP_ moad
11 gi ref YP_ XOO1089 ISXo8 transposase E-06 down YP_ gi ref YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO1158 XOO1159 XOO1160 XOO1161 fimbrial assembly membrane protein E-177 down YP_ piln fimbrial assembly membrane protein E-88 down YP_ pilo fimbrial assembly protein E-91 down YP_ pilp fimbrial assembly protein down YP_ pilq gi ref YP_ XOO1206 transposase E-126 up YP_ gi ref YP_ XOO1217 hypothetical protein down YP_ gi ref YP_ XOO1273 gi ref YP_ XOO1274 quinol oxidase subunit I E-91 down YP_ quinol oxidase subunit II E-72 down YP_ qxtb gi ref YP_ XOO1315 transposase E-168 up YP_ gi ref YP_ XOO1320 transposase E-161 up YP_ gi ref YP_ XOO1356 transporter up YP_ ycee gi ref YP_ XOO1380 HrpX protein E-76 down YP_ hrpxct gi ref YP_ XOO1392 IS1404 transposase E-07 up YP_ gi ref YP_ XOO1393 transposase E-133 up YP_ gi ref YP_ XOO1399 IS1479 transposase E-08 down YP_ gi ref YP_ XOO1413 peptidase up YP_ gi ref YP_ XOO1444 transposase E-132 up YP_ gi ref YP_ XOO1459 endonuclease V E-09 up YP_ nfi gi ref YP_ XOO1465 gi ref YP_ XOO1466 chemotaxis-specific methylesterase E-104 down YP_ cheb response regulator for chemotaxis E-82 down YP_ cher gi ref YP_ XOO1467 response regulator E-149 down YP_ viea gi ref YP_ XOO1468 chemotaxis protein down YP_ chew gi ref YP_ XOO1469 chemotaxis protein down YP_ mcp gi ref YP_ XOO1470 chemotaxis histidine protein kinase down YP_ chea gi ref YP_ XOO1471 hypothetical protein E-55 down YP_ gi ref YP_ XOO1481 hypothetical protein up YP_ cred gi ref YP_ XOO1487 cysteine protease E-150 down YP_ gi ref YP_ XOO1515 hypothetical protein up YP_ gi ref YP_ XOO1551 hypothetical protein E-37 down YP_ gi ref YP_ XOO1586 pilin E-276 down YP_ pile gi ref YP_ XOO1680 phage-related integrase up YP_ int gi ref YP_ XOO1681 hypothetical protein E-305 up YP_ gi ref YP_ XOO1682 hypothetical protein up YP_ gi ref YP_ XOO1683 hypothetical protein up YP_ gi ref YP_ XOO1684 hypothetical protein E-102 up YP_
12 gi ref YP_ XOO1685 hypothetical protein up YP_ gi ref YP_ XOO1686 hypothetical protein up YP_ gi ref YP_ XOO1687 hypothetical protein up YP_ gi ref YP_ XOO1688 hypothetical protein E-28 up YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO1689 XOO1690 XOO1691 phage-related tail protein up YP_ phage-related tail protein E-183 up YP_ phage-related tail protein up YP_ gi ref YP_ XOO1692 hypothetical protein E-179 up YP_ gi ref YP_ gi ref YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO1693 XOO1694 XOO1695 XOO1696 XOO1697 phage-related tail protein up YP_ phage-related tail protein up YP_ phage-related tail protein up YP_ phage-related baseplate protein up YP_ phage-related baseplate protein up YP_ gi ref YP_ XOO1698 hypothetical protein up YP_ gi ref YP_ gi ref YP_ XOO1699 XOO1700 phage-related tail protein up YP_ phage-related baseplate assembly protein up YP_ gi ref YP_ XOO1701 hypothetical protein up YP_ gi ref YP_ gi ref YP_ XOO1702 XOO1703 phage-related tail protein up YP_ phage-related tail protein up YP_ gi ref YP_ XOO1704 hypothetical protein up YP_ gi ref YP_ XOO1705 phage-related lytic enzyme up YP_ lys gi ref YP_ XOO1706 hypothetical protein up YP_ gi ref YP_ XOO1707 hypothetical protein up YP_ gi ref YP_ XOO1708 phage-related tail protein E-144 up YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO1709 XOO1710 XOO1712 phage-related capsid completion protein up YP_ phage-related terminase up YP_ phage-related major capsid protein up YP_ gi ref YP_ gi ref YP_ XOO1713 XOO1714 phage-related capsid scaffold protein up YP_ phage-related terminase up YP_ gi ref YP_ XOO1715 gi ref YP_ XOO1717 phage-related capsid packaging protein up YP_ site-specific DNAmethyltransferase up YP_ gi ref YP_ XOO1718 hypothetical protein E-67 up YP_ gi ref YP_ XOO1726 transposase E-199 up YP_ gi ref YP_ XOO1742 hypothetical protein E-08 up YP_
13 gi ref YP_ XOO1746 pilus protein down YP_ pilg gi ref YP_ XOO1747 PilH protein E-50 down YP_ pilh gi ref YP_ gi ref YP_ XOO1748 XOO1749 pilus biogenesis protein E-101 down YP_ pili pilus biogenesis protein E-176 down YP_ pilj gi ref YP_ gi ref YP_ XOO1754 XOO1784 ribosomal protein alanine acetyltransferase E-09 down YP_ rimj TonB-dependent receptor down YP_ iron gi ref YP_ XOO1864 polyketide synthase up YP_ frne gi ref YP_ XOO1887 hypothetical protein down YP_ gi ref YP_ XOO1949 transposase E-38 up YP_ gi ref YP_ gi ref YP_ XOO1978 XOO1981 pili assembly chaperone E-28 down YP_ pili assembly chaperone E-58 down YP_ ecpd gi ref YP_ XOO1982 protein U E-51 down YP_ pru gi ref YP_ XOO1998 MarR family transcriptional regulator E-09 up YP_ yyba gi ref YP_ XOO2010 transposase up YP_ gi ref YP_ XOO2011 transposase E-117 up YP_ gi ref YP_ XOO2087 transposase E-33 up YP_ gi ref YP_ XOO2088 transposase E-124 up YP_ gi ref YP_ XOO2100 transposase E-109 up YP_ gi ref YP_ XOO2102 ISXo8 transposase down YP_ gi ref YP_ XOO2107 transposase E-112 up YP_ gi ref YP_ XOO2158 transposase E-160 up YP_ gi ref YP_ XOO2203 hypothetical protein down YP_ gi ref YP_ gi ref YP_ XOO2204 XOO2227 methyl-accepting chemotaxis protein down YP_ tlpc two-component system regulatory protein E-24 down YP_ regr gi ref YP_ XOO2235 hypothetical protein up YP_ gi ref YP_ XOO2285 transposase E-129 up YP_ gi ref YP_ XOO2301 transposase E-127 up YP_ gi ref YP_ XOO2302 hypothetical protein E-15 up YP_ gi ref YP_ XOO2348 hypothetical protein E-69 down YP_ gi ref YP_ XOO2354 hypothetical protein E-236 down YP_ gi ref YP_ gi ref YP_ XOO2377 XOO2479 cation efflux system protein up YP_ czca site-specific recombinase up YP_ gi ref YP_ XOO2516 transposase E-194 up YP_ gi ref YP_ XOO2558 chemotaxis protein down YP_ mcp gi ref YP_ XOO2559 hypothetical protein E-21 down YP_
14 gi ref YP_ XOO2561 hypothetical protein E-101 down YP_ gi ref YP_ XOO2565 hypothetical protein E-285 down YP_ gi ref YP_ XOO2566 flagellar protein down YP_ flgm gi ref YP_ XOO2567 flagellar basal body P-ring biosynthesis protein FlgA E-149 down YP_ flga gi ref YP_ XOO2568 chemotaxis protein E-159 down YP_ chev gi ref YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO2569 XOO2570 XOO2571 XOO2572 gi ref YP_ XOO2574 gi ref YP_ XOO2575 gi ref YP_ XOO2576 flagellar basal-body rod protein FlgB E-90 down YP_ flgb flagellar basal body rod protein FlgC E-101 down YP_ flgc flagellar basal body rod modification protein E-221 down YP_ flgd flagellar hook protein FlgE down YP_ flge flagellar basal body rod protein FlgF E-140 down YP_ flgf flagellar basal body rod protein FlgG E-264 down YP_ flgg flagellar basal body L-ring protein E-233 down YP_ flgh gi ref YP_ gi ref YP_ gi ref YP_ XOO2577 XOO2578 XOO2579 flagellar basal body P-ring biosynthesis protein FlgA E-163 down YP_ flgi flagellar rod assembly protein/muramidase FlgJ E-113 down YP_ flgj flagellar hookassociated protein FlgK E-115 down YP_ flgk gi ref YP_ XOO2580 flagellar hookassociated protein FlgL E-121 down YP_ flgl gi ref YP_ XOO2581 flagellin down YP_ flic gi ref YP_ XOO2582 flagellar protein down YP_ flid gi ref YP_ XOO2583 flagellar protein E-113 down YP_ flis gi ref YP_ XOO2584 hypothetical protein E-130 down YP_ gi ref YP_ XOO2585 hypothetical protein E-31 down YP_ gi ref YP_ XOO2587 RNA polymerase sigma-54 factor E-267 down YP_ rpon gi ref YP_ XOO2588 response regulator E-43 down YP_ gi ref YP_ XOO2591 acyl carrier protein E-17 down YP_ acp gi ref YP_ gi ref YP_ XOO2592 XOO oxoacyl-ACP synthase E-56 down YP_ fabh oxoacyl-ACP reductase E-17 down YP_ fabg gi ref YP_ XOO2594 dehydrogenase E-29 down YP_ gi ref YP_ XOO2596 ring hydroxylating dioxygenase subunit alpha E-39 down YP_ gi ref YP_ XOO2598 hypothetical protein E-38 down YP_ gi ref YP_ XOO2600 flagellar protein E-30 down YP_ flie gi ref YP_ XOO2601 flagellar MS-ring protein E-242 down YP_ flif gi ref YP_ XOO2602 flagellar protein E-96 down YP_ flig
15 gi ref YP_ XOO2604 flagellar protein E-34 down YP_ flii gi ref YP_ XOO2607 flagellar protein E-62 down YP_ flil gi ref YP_ XOO2608 flagellar motor switch protein FliM E-34 down YP_ flim gi ref YP_ XOO2609 flagellar protein E-13 down YP_ flin gi ref YP_ XOO2610 flagellar protein E-19 down YP_ flio gi ref YP_ gi ref YP_ gi ref YP_ XOO2611 XOO2612 XOO2613 flagellar biosynthesis protein FliP E-19 down YP_ flip flagellar biosynthesis E-22 down YP_ fliq flagellar biosynthetic protein E-15 down YP_ flir gi ref YP_ XOO2614 diguanylate cyclase E-43 down YP_ gi ref YP_ XOO2616 diguanylate cyclase E-11 down YP_ gi ref YP_ XOO2617 flagellar biosynthesis protein FlhB E-65 down YP_ flhb gi ref YP_ XOO2618 flagellar biosynthesis protein FlhA E-69 down YP_ flha gi ref YP_ XOO2619 gi ref YP_ gi ref YP_ XOO2620 XOO2621 flagellar biosynthesis regulator FlhF E-157 down YP_ flhf flagellar biosynthesis switch protein E-108 down YP_ flen RNA polymerase sigma factor FliA down YP_ flia gi ref YP_ XOO2622 chemotaxis protein E-216 down YP_ chey gi ref YP_ gi ref YP_ XOO2623 XOO2624 chemotaxis related protein E-155 down YP_ chez chemotaxis related protein down YP_ chea gi ref YP_ XOO2642 transporter E-07 up YP_ acrd gi ref YP_ XOO2648 transposase E-140 up YP_ gi ref YP_ XOO2661 cytochrome D ubiquinol oxidase subunit II up YP_ cydb gi ref YP_ XOO2738 virulence protein E-11 down YP_ acvb gi ref YP_ XOO2749 transposase E-202 up YP_ gi ref YP_ XOO2782 glutamine synthetase E-34 down YP_ glna gi ref YP_ XOO2787 transcriptional regulator E-238 down YP_ rrpx gi ref YP_ XOO2807 transposase E-137 up YP_ gi ref YP_ gi ref YP_ gi ref YP_ XOO2830 XOO2831 XOO2832 flagellar motor protein E-189 down YP_ motc flagellar motor protein MotD E-91 down YP_ motb chromosome partioning protein E-144 down YP_ para gi ref YP_ XOO2833 chemotaxis protein down YP_ chew gi ref YP_ XOO2834 hypothetical protein E-197 down YP_ gi ref YP_ XOO2835 chemotaxis response regulator E-140 down YP_ chey gi ref YP_ XOO2836 chemotaxis protein down YP_ chea
16 gi ref YP_ XOO2840 chemotaxis protein E-115 down YP_ tsr gi ref YP_ XOO2841 transposase E-156 up YP_ gi ref YP_ XOO2842 chemotaxis protein down YP_ tsr gi ref YP_ XOO2844 chemotaxis protein down YP_ tsr gi ref YP_ XOO2847 chemotaxis protein down YP_ tsr gi ref YP_ XOO2848 chemotaxis protein down YP_ tsr gi ref YP_ XOO2849 hypothetical protein E-52 down YP_ gi ref YP_ XOO2850 chemotaxis protein E-136 down YP_ chew gi ref YP_ XOO2857 chemotaxis protein methyltransferase E-202 down YP_ cher gi ref YP_ XOO2883 transposase E-134 up YP_ gi ref YP_ XOO2940 recombination regulator RecX E-100 up YP_ recx gi ref YP_ XOO2941 recombinase A up YP_ reca gi ref YP_ XOO2981 ABC transporter ATP-binding protein E-06 up YP_ gi ref YP_ XOO3000 cytochrome C up YP_ gi ref YP_ XOO3012 hypothetical protein up YP_ gi ref YP_ XOO3095 transposase E-53 up YP_ gi ref YP_ XOO3097 transposase E-121 up YP_ gi ref YP_ XOO3109 transposase E-164 up YP_ gi ref YP_ XOO3125 peptidase E-12 up YP_ gi ref YP_ XOO3126 hypothetical protein E-12 down YP_ gi ref YP_ XOO3195 type IV pilin E-30 up YP_ pile gi ref YP_ XOO3196 protein PilY up YP_ pily gi ref YP_ XOO3197 transposase E-299 up YP_ gi ref YP_ XOO3199 protein PilX E-73 down YP_ pilx gi ref YP_ XOO3200 hypothetical protein down YP_ gi ref YP_ gi ref YP_ XOO3201 XOO3202 pre-pilin leader sequence E-95 down YP_ pilv pre-pilin like leader sequence E-131 down YP_ fimt gi ref YP_ XOO3207 hypothetical protein down YP_ gi ref YP_ XOO3246 phosphodiesterase E-69 down YP_ gi ref YP_ XOO3247 hypothetical protein E-104 down YP_ gi ref YP_ XOO3268 hypothetical protein down YP_ gi ref YP_ XOO3284 IS1595 transposase E-45 up YP_ gi ref YP_ XOO3300 extracellular protease up YP_ gi ref YP_ XOO3345 hypothetical protein E-43 down YP_ gi ref YP_ XOO3346 transposase E-106 up YP_ gi ref YP_ XOO3353 transposase E-10 up YP_
17 gi ref YP_ XOO3354 transposase E-25 up YP_ gi ref YP_ XOO3364 hypothetical protein E-29 down YP_ gi ref YP_ XOO3375 hypothetical protein E-15 down YP_ gi ref YP_ XOO3380 gi ref YP_ gi ref YP_ XOO3381 XOO3388 fumarylacetoacetate hydrolase E-49 up YP_ large-conductance mechanosensitive channel E-220 up YP_ mscl trna/rrna methyltransferase E-50 up YP_ gi ref YP_ XOO3391 transposase E-198 up YP_ gi ref YP_ XOO3392 hypothetical protein E-08 up YP_ gi ref YP_ XOO3393 aminopeptidase E-50 up YP_ pepn gi ref YP_ XOO3394 acriflavin resistance protein E-63 up YP_ acrf gi ref YP_ XOO3395 RND efflux membrane fusion protein E-30 up YP_ gi ref YP_ XOO3396 bifunctional sulfate adenylyltransferase subunit 1/adenylylsulfate kinase E-86 up YP_ nodq gi ref YP_ XOO3397 gi ref YP_ XOO3400 gi ref YP_ XOO3401 sulfate adenylyltransferase subunit E-57 up YP_ raxp sulfite reductase subunit beta E-21 up YP_ cysi phosphoadenosine phosphosulfate reductase E-10 up YP_ cysh gi ref YP_ XOO3612 hypothetical protein E-111 down YP_ gi ref YP_ gi ref YP_ XOO3646 XOO3647 alkyl hydroperoxide reductase E-78 down YP_ ahpf oxidative stress transcriptional regulator E-66 down YP_ oxyr gi ref YP_ XOO3725 hypothetical protein E-22 up YP_ gi ref YP_ XOO3737 transposase E-143 up YP_ gi ref YP_ gi ref YP_ XOO3751 XOO3846 dihydrofolate reductase type III E-21 up YP_ fola potassiumtransporting ATPase subunit A up YP_ kdpa gi ref YP_ XOO3910 transposase E-194 up YP_ gi ref YP_ XOO3935 two-component system regulatory protein E-28 down YP_ gi ref YP_ XOO3988 response regulator E-142 down YP_ gi ref YP_ XOO3989 aminopeptidase up YP_ pepn gi ref YP_ XOO4003 hypothetical protein E-28 up YP_ gi ref YP_ XOO4008 two-component system regulatory protein E-48 down YP_ algr gi ref YP_ XOO4021 histidine kinaseresponse regulator hybrid protein E-121 down YP_ gi ref YP_ XOO4030 trehalase E-47 down YP_ trea
18 gi ref YP_ XOO4048 threonine aldolase up YP_ gi ref YP_ XOO4076 nuclease E-08 up YP_ nuch gi ref YP_ XOO4077 Pyruvate dehydrogenase e1 component subunit alpha up YP_ pdha gi ref YP_ XOO4079 transposase E-160 up YP_ gi ref YP_ XOO4081 Pyruvate dehydrogenase E1 component subunit beta E-30 up YP_ pdhb gi ref YP_ XOO4082 hypothetical protein up YP_ gi ref YP_ XOO4097 hypothetical protein down YP_ gi ref YP_ XOO4105 transposase E-234 up YP_ gi ref YP_ XOO4112 hypothetical protein up YP_ gi ref YP_ XOO4117 transposase E-237 up YP_ gi ref YP_ XOO4118 transposase E-13 up YP_ gi ref YP_ XOO4121 transposase E-210 up YP_ gi ref YP_ XOO4147 transposase E-118 up YP_ gi ref YP_ XOO4150 bacterioferritinassociated ferredoxin down YP_ gi ref YP_ XOO4193 transposase E-150 up YP_ gi ref YP_ XOO4204 hypothetical protein E-08 up YP_ gi ref YP_ XOO4217 hypothetical protein up YP_ gi ref YP_ XOO4220 hypothetical protein E-65 down YP_ gi ref YP_ XOO4223 transposase up YP_ gi ref YP_ XOO4224 transposase E-133 up YP_ gi ref YP_ XOO4275 hypothetical protein up YP_ gi ref YP_ XOO4281 transposase E-25 up YP_ gi ref YP_ XOO4287 hypothetical protein E-30 down YP_ gi ref YP_ XOO methyltetrahydropte royltriglutamate/ho mocysteine S- methyltransferase down YP_ mete gi ref YP_ XOO4334 hypothetical protein down YP_ gi ref YP_ XOO4335 NADH-dependent FMN reductase E-102 down YP_ sfla gi ref YP_ XOO4356 alpha/beta hydrolase E-09 up YP_ gi ref YP_ XOO4357 transposase E-121 up YP_ gi ref YP_ XOO4431 TonB-dependent receptor E-15 down YP_ cira gi ref YP_ XOO4440 transposase E-134 up YP_ gi ref YP_ XOO4441 transposase E-21 up YP_ gi ref YP_ XOO4442 hypothetical protein up YP_
19 gi ref YP_ XOO4472 hypothetical protein E-39 down YP_ gi ref YP_ XOO4479 transposase E-164 up YP_ gi ref YP_ XOO4520 transposase E-161 up YP_ gi ref YP_ XOO4571 transposase E-185 up YP_ gi ref YP_ XOO4580 hypothetical protein E-14 up YP_ gi ref YP_ XOO4614 hypothetical protein E-28 up YP_ gi ref YP_ XOO4615 hypothetical protein up YP_ gi ref YP_ XOO4616 IS1478 transposase E-58 down YP_ gi ref YP_ XOO4703 transposase E-14 up gi ref YP_ XOO4734 transposase E-101 up gi ref YP_ XOO4779 hypothetical protein up gi ref YP_ XOO4782 transposase E-18 up gi ref YP_ XOO4790 hypothetical protein E-06 up gi ref YP_ XOO4825 hypothetical protein E-26 down gi ref YP_ XOO4876 transposase E-08 up Table S2. DEGs between WT and ΔdgcA mutant. YP_ YP_ YP_ YP_ YP_ YP_ YP_
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Supporting Information
 Supporting Information Mitochondria-Targeting Polydopamine Nanocomposites as Chemophotothermal Therapeutics for Cancer Zhuo Wang *,, Yuzhi Chen, Hui Zhang, Yawen Li, Yufan Ma, Jia Huang, Xiaolei Liu, Fang
Supporting Information Mitochondria-Targeting Polydopamine Nanocomposites as Chemophotothermal Therapeutics for Cancer Zhuo Wang *,, Yuzhi Chen, Hui Zhang, Yawen Li, Yufan Ma, Jia Huang, Xiaolei Liu, Fang
Quantum dot sensitized solar cells with efficiency over 12% based on tetraethyl orthosilicate additive in polysulfide electrolyte
 Electronic Supplementary Material (ESI) for Journal of Materials Chemistry A. This journal is The Royal Society of Chemistry 2017 Supplementary Information (SI) Quantum dot sensitized solar cells with
Electronic Supplementary Material (ESI) for Journal of Materials Chemistry A. This journal is The Royal Society of Chemistry 2017 Supplementary Information (SI) Quantum dot sensitized solar cells with
Acknowledgements... 3 Contents... I Figures... VII Tables... IX Abbreviations... X
 I Acknowledgements... 3... I Figures... VII Tables... IX Abbreviations... X Amino acids... XII Nucleobases... XII 1 Introduction... 1 1.1 Polyketides and non-ribosomal peptides... 1 1.2 Polyketide synthases...
I Acknowledgements... 3... I Figures... VII Tables... IX Abbreviations... X Amino acids... XII Nucleobases... XII 1 Introduction... 1 1.1 Polyketides and non-ribosomal peptides... 1 1.2 Polyketide synthases...
JCVI Role Category_1 JCVI Role Category_2. histidinol phosphatase-related protein Unknown function NT01ST0303 AAL19211 STM0248
 Additional Table 3. Detailed information on all 81 genes that showed evidence for positive selection from initial positive selection analysis, in Protein Name Gene name JCVI Role Category_1 JCVI Role Category_2
Additional Table 3. Detailed information on all 81 genes that showed evidence for positive selection from initial positive selection analysis, in Protein Name Gene name JCVI Role Category_1 JCVI Role Category_2
D-Glucosamine-derived copper catalyst for Ullmann-type C- N coupling reaction: theoretical and experimental study
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 D-Glucosamine-derived copper catalyst for Ullmann-type C- N coupling reaction: theoretical
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 D-Glucosamine-derived copper catalyst for Ullmann-type C- N coupling reaction: theoretical
Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic acids with the assistance of isocyanide
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
Supporting Information. Asymmetric Binary-acid Catalysis with Chiral. Phosphoric Acid and MgF 2 : Catalytic
 Supporting Information Asymmetric Binary-acid Catalysis with Chiral Phosphoric Acid and MgF 2 : Catalytic Enantioselective Friedel-Crafts Reactions of β,γ- Unsaturated-α-Ketoesters Jian Lv, Xin Li, Long
Supporting Information Asymmetric Binary-acid Catalysis with Chiral Phosphoric Acid and MgF 2 : Catalytic Enantioselective Friedel-Crafts Reactions of β,γ- Unsaturated-α-Ketoesters Jian Lv, Xin Li, Long
JCVI Locus Protein Name. JCVI Role Category JCVI Locus (LT2) Annotation name
 Additional File 4. Detailed information on all 41 genes that showed evidence for positive selection from positive selection analysis, in which gene Alignment Primary Gene Genbank Locus JCVI Locus Protein
Additional File 4. Detailed information on all 41 genes that showed evidence for positive selection from positive selection analysis, in which gene Alignment Primary Gene Genbank Locus JCVI Locus Protein
Electronic Supplementary Information
 Electronic Supplementary Information Unprecedented Carbon-Carbon Bond Cleavage in Nucleophilic Aziridine Ring Opening Reaction, Efficient Ring Transformation of Aziridines to Imidazolidin-4-ones Jin-Yuan
Electronic Supplementary Information Unprecedented Carbon-Carbon Bond Cleavage in Nucleophilic Aziridine Ring Opening Reaction, Efficient Ring Transformation of Aziridines to Imidazolidin-4-ones Jin-Yuan
Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo
 Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
Copper-Catalyzed Oxidative Dehydrogenative N-N Bond. Formation for the Synthesis of N,N -Diarylindazol-3-ones
 Electronic Supplementary Material (ESI) for Organic Chemistry Frontiers. This journal is the Partner Organisations 2016 Supporting information Copper-Catalyzed Oxidative Dehydrogenative - Bond Formation
Electronic Supplementary Material (ESI) for Organic Chemistry Frontiers. This journal is the Partner Organisations 2016 Supporting information Copper-Catalyzed Oxidative Dehydrogenative - Bond Formation
Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance
 POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
College of Life Science, Dalian Nationalities University, Dalian , PR China.
 Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2018 Postsynthetic modification
Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2018 Postsynthetic modification
(1) A lecturer at the University College of Applied Sciences in Gaza. Gaza, Palestine, P.O. Box (1514).
 1439 2017, 3,29, 1658 7677: 1439 2017,323 299, 3,29, 1 1438 09 06 ; 1438 02 23 : 33,,,,,,,,,,, : The attitudes of lecturers at the University College of Applied Sciences in Gaza (BA - Diploma) towards
1439 2017, 3,29, 1658 7677: 1439 2017,323 299, 3,29, 1 1438 09 06 ; 1438 02 23 : 33,,,,,,,,,,, : The attitudes of lecturers at the University College of Applied Sciences in Gaza (BA - Diploma) towards
Supplementary Information. Living Ring-Opening Polymerization of Lactones by N-Heterocyclic Olefin/Al(C 6 F 5 ) 3
 Supplementary Information Living Ring-Opening Polymerization of Lactones by N-Heterocyclic Olefin/Al(C 6 F 5 ) 3 Lewis Pairs: Structures of Intermediates, Kinetics, and Mechanism Qianyi Wang, Wuchao Zhao,
Supplementary Information Living Ring-Opening Polymerization of Lactones by N-Heterocyclic Olefin/Al(C 6 F 5 ) 3 Lewis Pairs: Structures of Intermediates, Kinetics, and Mechanism Qianyi Wang, Wuchao Zhao,
Table S1. Summary of data collections and structure refinements for crystals 1Rb-1h, 1Rb-2h, and 1Rb-4h.
 Supporting Information [NH 3 CH 3 ] [In SbS 9 SH]: A novel methylamine-directed indium thioantimonate with Rb + ion-exchange property Kai-Yao Wang a,b, Mei-Ling Feng a, Jian-Rong Li a and Xiao-Ying Huang
Supporting Information [NH 3 CH 3 ] [In SbS 9 SH]: A novel methylamine-directed indium thioantimonate with Rb + ion-exchange property Kai-Yao Wang a,b, Mei-Ling Feng a, Jian-Rong Li a and Xiao-Ying Huang
Aluminum Electrolytic Capacitors (Large Can Type)
 Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Computational study of the structure, UV-vis absorption spectra and conductivity of biphenylene-based polymers and their boron nitride analogues
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Computational study of the structure, UV-vis absorption spectra and conductivity of biphenylene-based
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Computational study of the structure, UV-vis absorption spectra and conductivity of biphenylene-based
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
Supporting Information. Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka
 Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supporting Information
 Supporting Information Lewis acid catalyzed ring-opening reactions of methylenecyclopropanes with diphenylphosphine oxide in the presence of sulfur or selenium Min Shi,* Min Jiang and Le-Ping Liu State
Supporting Information Lewis acid catalyzed ring-opening reactions of methylenecyclopropanes with diphenylphosphine oxide in the presence of sulfur or selenium Min Shi,* Min Jiang and Le-Ping Liu State
Potential Dividers. 46 minutes. 46 marks. Page 1 of 11
 Potential Dividers 46 minutes 46 marks Page 1 of 11 Q1. In the circuit shown in the figure below, the battery, of negligible internal resistance, has an emf of 30 V. The pd across the lamp is 6.0 V and
Potential Dividers 46 minutes 46 marks Page 1 of 11 Q1. In the circuit shown in the figure below, the battery, of negligible internal resistance, has an emf of 30 V. The pd across the lamp is 6.0 V and
Comparison of carbon-sulfur and carbon-amine bond in therapeutic drug: -S-aromatic heterocyclic podophyllum derivatives display antitumor activity
 Comparison of carbon-sulfur and carbon-amine bond in therapeutic drug: -S-aromatic heterocyclic podophyllum derivatives display antitumor activity Jian-Long Li 1,a, Wei Zhao 1,a, Chen Zhou 1,a, Ya-Xuan
Comparison of carbon-sulfur and carbon-amine bond in therapeutic drug: -S-aromatic heterocyclic podophyllum derivatives display antitumor activity Jian-Long Li 1,a, Wei Zhao 1,a, Chen Zhou 1,a, Ya-Xuan
the total number of electrons passing through the lamp.
 1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil
 J. Jpn. Soc. Soil Phys. No. +*0, p.- +*,**1 Eh * ** Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil Daisuke MURAKAMI* and Tatsuaki KASUBUCHI** * The United Graduate
J. Jpn. Soc. Soil Phys. No. +*0, p.- +*,**1 Eh * ** Reaction of a Platinum Electrode for the Measurement of Redox Potential of Paddy Soil Daisuke MURAKAMI* and Tatsuaki KASUBUCHI** * The United Graduate
Η ΨΥΧΙΑΤΡΙΚΗ - ΨΥΧΟΛΟΓΙΚΗ ΠΡΑΓΜΑΤΟΓΝΩΜΟΣΥΝΗ ΣΤΗΝ ΠΟΙΝΙΚΗ ΔΙΚΗ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΝΟΜΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΤΟΜΕΑΣ ΙΣΤΟΡΙΑΣ ΦΙΛΟΣΟΦΙΑΣ ΚΑΙ ΚΟΙΝΩΝΙΟΛΟΓΙΑΣ ΤΟΥ ΔΙΚΑΙΟΥ Διπλωματική εργασία στο μάθημα «ΚΟΙΝΩΝΙΟΛΟΓΙΑ ΤΟΥ ΔΙΚΑΙΟΥ»
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΝΟΜΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΤΟΜΕΑΣ ΙΣΤΟΡΙΑΣ ΦΙΛΟΣΟΦΙΑΣ ΚΑΙ ΚΟΙΝΩΝΙΟΛΟΓΙΑΣ ΤΟΥ ΔΙΚΑΙΟΥ Διπλωματική εργασία στο μάθημα «ΚΟΙΝΩΝΙΟΛΟΓΙΑ ΤΟΥ ΔΙΚΑΙΟΥ»
Σχέση µεταξύ της Μεθόδου των ερµατοπτυχών και της Βιοηλεκτρικής Αντίστασης στον Υπολογισµό του Ποσοστού Σωµατικού Λίπους
 Ερευνητική Aναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµότόµος 1(3), 244 251 ηµοσιεύτηκε: 2 8 εκεµβρίου 2003 Inquiries in Sport & Physical Education Volume 1(3), 244 251 Released: December 28, 2003 www.hape.gr/emag.asp
Ερευνητική Aναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµότόµος 1(3), 244 251 ηµοσιεύτηκε: 2 8 εκεµβρίου 2003 Inquiries in Sport & Physical Education Volume 1(3), 244 251 Released: December 28, 2003 www.hape.gr/emag.asp
Identification of Fish Species using DNA Method
 5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
Free Radical Initiated Coupling Reaction of Alcohols and. Alkynes: not C-O but C-C Bond Formation. Context. General information 2. Typical procedure 2
 Free Radical Initiated Coupling Reaction of Alcohols and Alkynes: not C-O but C-C Bond Formation Zhongquan Liu,* Liang Sun, Jianguo Wang, Jie Han, Yankai Zhao, Bo Zhou Institute of Organic Chemistry, Gannan
Free Radical Initiated Coupling Reaction of Alcohols and Alkynes: not C-O but C-C Bond Formation Zhongquan Liu,* Liang Sun, Jianguo Wang, Jie Han, Yankai Zhao, Bo Zhou Institute of Organic Chemistry, Gannan
[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,
![[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13, [11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,](/thumbs/89/98694254.jpg) in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ Πρόγραμμα Μεταπτυχιακών Σπουδών του Τμήματος Bioyiiudac & Βιοτετνολονίας «Βιοτεχνολογία - Ποιότητα Διατροφής & Περιβάλλοντος» υγγιατ'1 '-ε,λχ-βε0τ Χ ϊο4ο Γ/4ΐ/ '****f*tf*wftieg{
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ Πρόγραμμα Μεταπτυχιακών Σπουδών του Τμήματος Bioyiiudac & Βιοτετνολονίας «Βιοτεχνολογία - Ποιότητα Διατροφής & Περιβάλλοντος» υγγιατ'1 '-ε,λχ-βε0τ Χ ϊο4ο Γ/4ΐ/ '****f*tf*wftieg{
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 651 APPENDIX B. BIBLIOGRAPHY 677 APPENDIX C. ANSWERS TO SELECTED EXERCISES 679
 APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
Optimizing Microwave-assisted Extraction Process for Paprika Red Pigments Using Response Surface Methodology
 2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
Technical Research Report, Earthquake Research Institute, the University of Tokyo, No. +-, pp. 0 +3,,**1. No ,**1
 No. +- 0 +3,**1 Technical Research Report, Earthquake Research Institute, the University of Tokyo, No. +-, pp. 0 +3,,**1. * Construction of the General Observation System for Strong Motion in Earthquake
No. +- 0 +3,**1 Technical Research Report, Earthquake Research Institute, the University of Tokyo, No. +-, pp. 0 +3,,**1. * Construction of the General Observation System for Strong Motion in Earthquake
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
Nguyen Hien Trang* **
 152 Nippon Shokuhin Kagaku Kogaku Kaishi Vol.., No.., +,+3 (,**1) 10 Nguyen Hien Trang* ** * Department of Food Science and Technology, Hue University of Agriculture and Forestry ** Properties of "Shishibishio
152 Nippon Shokuhin Kagaku Kogaku Kaishi Vol.., No.., +,+3 (,**1) 10 Nguyen Hien Trang* ** * Department of Food Science and Technology, Hue University of Agriculture and Forestry ** Properties of "Shishibishio
Science of Sericulture
 Science of Sericulture 2012 38 1 0065-0069 ISSN 0257-4799 CN 32-1115 /S E-mail CYKE@ chinajournal. net. cn 215123 25 0 ~ 24 h GSH GSSG GSH + 2GSSG GSH /GSSG S- GST TPX 23% GR 57% GSH TPX GST GSSG 61% GSH
Science of Sericulture 2012 38 1 0065-0069 ISSN 0257-4799 CN 32-1115 /S E-mail CYKE@ chinajournal. net. cn 215123 25 0 ~ 24 h GSH GSSG GSH + 2GSSG GSH /GSSG S- GST TPX 23% GR 57% GSH TPX GST GSSG 61% GSH
2 0 1 2 2 Journal of Shanghai Normal University Natural Sciences Feb. 2 0 1 2 SNAT2. HEK293T Western blot SNAT2 - HA Q 31 A 1000-5137 2012 01-0083-06
 4 1 1 Vol 41 No 1 2 0 1 2 2 Journal of Shanghai Normal University Natural Sciences Feb 2 0 1 2 SNAT2 - HA * 200234 SNAT2 - SNAT2 PCR HA SNAT2 C pbk - CMVΔ 1098-1300 - SNAT2 - HA HEK293T Western blot SNAT2
4 1 1 Vol 41 No 1 2 0 1 2 2 Journal of Shanghai Normal University Natural Sciences Feb 2 0 1 2 SNAT2 - HA * 200234 SNAT2 - SNAT2 PCR HA SNAT2 C pbk - CMVΔ 1098-1300 - SNAT2 - HA HEK293T Western blot SNAT2
Aluminum Electrolytic Capacitors
 Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Microcosm Setup and Biomass Sampling. Seawater for microcosm incubation. August 16, 2007 during the CMORE Bloomer Cruise. Hydrocasts for sampling were
 SUPPORTING INFORMATION SUPPORTING TEXT METHODS Microcosm Setup and Biomass Sampling. Seawater for microcosm incubation experiments was collected at 23 12.88 N, 159 8.17 W, from 75-m depth, pre-dawn on
SUPPORTING INFORMATION SUPPORTING TEXT METHODS Microcosm Setup and Biomass Sampling. Seawater for microcosm incubation experiments was collected at 23 12.88 N, 159 8.17 W, from 75-m depth, pre-dawn on
Supplementary information:
 Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια (MIS 365280)
 «Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
«Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Supplementary Information for
 Supplentary Information for Aggregation induced blue-shifted ission molecular picture from QM/MM study Qunyan Wu, a Tian Zhang, a Qian Peng,* b Dong Wang, a and Zhigang Shuai* a a Key Loratory of Organic
Supplentary Information for Aggregation induced blue-shifted ission molecular picture from QM/MM study Qunyan Wu, a Tian Zhang, a Qian Peng,* b Dong Wang, a and Zhigang Shuai* a a Key Loratory of Organic
The toxicity of three chitin synthesis inhibitors to Calliptamus italicus Othoptera Acridoidea
 2011 48 4 909 914 * 1 2** 2 2 2 2 2*** 1. 110161 2. 100081 026000 3 Calliptamus italicus L. 3 LC 50 LC 90 1. 34 14. 17 mg / L LC 50 LC 90 2. 09 45. 22 mg / L 50 mg / L 14 d 87% 100% 50 mg / L 50% The toxicity
2011 48 4 909 914 * 1 2** 2 2 2 2 2*** 1. 110161 2. 100081 026000 3 Calliptamus italicus L. 3 LC 50 LC 90 1. 34 14. 17 mg / L LC 50 LC 90 2. 09 45. 22 mg / L 50 mg / L 14 d 87% 100% 50 mg / L 50% The toxicity
Thin Film Chip Resistors
 FEATURES PRECISE TOLERANCE AND TEMPERATURE COEFFICIENT EIA STANDARD CASE SIZES (0201 ~ 2512) LOW NOISE, THIN FILM (NiCr) CONSTRUCTION REFLOW SOLDERABLE (Pb FREE TERMINATION FINISH) Type Size EIA PowerRating
FEATURES PRECISE TOLERANCE AND TEMPERATURE COEFFICIENT EIA STANDARD CASE SIZES (0201 ~ 2512) LOW NOISE, THIN FILM (NiCr) CONSTRUCTION REFLOW SOLDERABLE (Pb FREE TERMINATION FINISH) Type Size EIA PowerRating
Study on Re-adhesion control by monitoring excessive angular momentum in electric railway traction
 () () Study on e-adhesion control by monitoring excessive angular momentum in electric railway traction Takafumi Hara, Student Member, Takafumi Koseki, Member, Yutaka Tsukinokizawa, Non-member Abstract
() () Study on e-adhesion control by monitoring excessive angular momentum in electric railway traction Takafumi Hara, Student Member, Takafumi Koseki, Member, Yutaka Tsukinokizawa, Non-member Abstract
2. Η Βαρύτητα Διαφόρων Γλυκαιμικών Δεικτών Νοσηλείας στην Λειτουργική Έκβαση του ΑΕΕ των Διαβητικών Ασθενών
 26 2. Η Βαρύτητα Διαφόρων Γλυκαιμικών Δεικτών Νοσηλείας στην Λειτουργική Έκβαση του ΑΕΕ των Διαβητικών Ασθενών Β. Δραγουμάνος 1, Δ. Αθανασόπουλος 2, Ε. Φουστέρης 1, Δ. Αναστασόπουλος 2, Π. Φουρλεμάδης
26 2. Η Βαρύτητα Διαφόρων Γλυκαιμικών Δεικτών Νοσηλείας στην Λειτουργική Έκβαση του ΑΕΕ των Διαβητικών Ασθενών Β. Δραγουμάνος 1, Δ. Αθανασόπουλος 2, Ε. Φουστέρης 1, Δ. Αναστασόπουλος 2, Π. Φουρλεμάδης
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ Πρόγραμμα Μεταπτυχιακών Σπουδών «Ολοκληρωμένη Ανάπτυξη & Διαχείριση Αγροτικού Χώρου» ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΑΤΡΙΒΗ «Η συμβολή των Τοπικών Προϊόντων
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ Πρόγραμμα Μεταπτυχιακών Σπουδών «Ολοκληρωμένη Ανάπτυξη & Διαχείριση Αγροτικού Χώρου» ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΑΤΡΙΒΗ «Η συμβολή των Τοπικών Προϊόντων
1 (forward modeling) 2 (data-driven modeling) e- Quest EnergyPlus DeST 1.1. {X t } ARMA. S.Sp. Pappas [4]
![1 (forward modeling) 2 (data-driven modeling) e- Quest EnergyPlus DeST 1.1. {X t } ARMA. S.Sp. Pappas [4] 1 (forward modeling) 2 (data-driven modeling) e- Quest EnergyPlus DeST 1.1. {X t } ARMA. S.Sp. Pappas [4]](/thumbs/91/106768733.jpg) 212 2 ( 4 252 ) No.2 in 212 (Total No.252 Vol.4) doi 1.3969/j.issn.1673-7237.212.2.16 STANDARD & TESTING 1 2 2 (1. 2184 2. 2184) CensusX12 ARMA ARMA TU111.19 A 1673-7237(212)2-55-5 Time Series Analysis
212 2 ( 4 252 ) No.2 in 212 (Total No.252 Vol.4) doi 1.3969/j.issn.1673-7237.212.2.16 STANDARD & TESTING 1 2 2 (1. 2184 2. 2184) CensusX12 ARMA ARMA TU111.19 A 1673-7237(212)2-55-5 Time Series Analysis
Electrolyzed-Reduced Water as Artificial Hot Spring Water
 /-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
/-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
Supporting Information. Generation Response. Physics & Chemistry of CAS, 40-1 South Beijing Road, Urumqi , China. China , USA
 Supporting Information Pb 3 B 6 O 11 F 2 : A First Noncentrocentric Lead Fluoroborate with Large Second Harmonic Generation Response Hongyi Li, a Hongping Wu, a * Xin Su, a Hongwei Yu, a,b Shilie Pan,
Supporting Information Pb 3 B 6 O 11 F 2 : A First Noncentrocentric Lead Fluoroborate with Large Second Harmonic Generation Response Hongyi Li, a Hongping Wu, a * Xin Su, a Hongwei Yu, a,b Shilie Pan,
Strain gauge and rosettes
 Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά
 ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ ΝΟΣΗΛΕΥΤΙΚΗ 2008, 47(3):367 373 H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά Ανθή Χρυσοστόμου Νοσηλεύτρια ΠΕ, ΜSc, Kέντρο Ελέγχου και Πρόληψης
ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ ΝΟΣΗΛΕΥΤΙΚΗ 2008, 47(3):367 373 H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά Ανθή Χρυσοστόμου Νοσηλεύτρια ΠΕ, ΜSc, Kέντρο Ελέγχου και Πρόληψης
Metal-free Oxidative Coupling of Amines with Sodium Sulfinates: A Mild Access to Sulfonamides
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting information for Metal-free Oxidative Coupling of Amines with Sodium Sulfinates:
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting information for Metal-free Oxidative Coupling of Amines with Sodium Sulfinates:
Dietary success of a new key fish in an overfished ecosystem: evidence from fatty acid and stable isotope signatures
 The following supplement accompanies the article Dietary success of a new key fish in an overfished ecosystem: evidence from fatty acid and stable isotope signatures M. G. van der Bank 1, A. C. Utne-Palm
The following supplement accompanies the article Dietary success of a new key fish in an overfished ecosystem: evidence from fatty acid and stable isotope signatures M. G. van der Bank 1, A. C. Utne-Palm
«Συντήρηση αχλαδιών σε νερό. υπό την παρουσία σπόρων σιναπιού (Sinapis arvensis).»
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΓΕΩΠΟΝΙΚΗ ΣΧΟΛΗ ΤΟΜΕΑΣ ΕΠΙΣΤΗΜΗΣ & ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΕΛΕΝΗ Π. ΠΑΠΑΤΣΑΡΟΥΧΑ Πτυχιούχος Τεχνολόγος Τροφίμων της Γεωπονικής Σχολής (Α.Π.Θ.) «Συντήρηση αχλαδιών σε
C H Activation of Cp* Ligand Coordinated to Ruthenium. Center: Synthesis and Reactivity of a Thiolate-Bridged
 Supporting Information C H Activation of Cp* Ligand Coordinated to Ruthenium Center: Synthesis and Reactivity of a Thiolate-Bridged Diruthenium Complex Featuring Fulvene-like Cp* Ligand Xiaoxiao Ji, Dawei
Supporting Information C H Activation of Cp* Ligand Coordinated to Ruthenium Center: Synthesis and Reactivity of a Thiolate-Bridged Diruthenium Complex Featuring Fulvene-like Cp* Ligand Xiaoxiao Ji, Dawei
( ) , ) , ; kg 1) 80 % kg. Vol. 28,No. 1 Jan.,2006 RESOURCES SCIENCE : (2006) ,2 ,,,, ; ;
 28 1 2006 1 RESOURCES SCIENCE Vol. 28 No. 1 Jan. 2006 :1007-7588(2006) 01-0002 - 07 20 1 1 2 (11 100101 ; 21 101149) : 1978 1978 2001 ; 2010 ; ; ; : ; ; 24718kg 1) 1990 26211kg 260kg 1995 2001 238kg( 1)
28 1 2006 1 RESOURCES SCIENCE Vol. 28 No. 1 Jan. 2006 :1007-7588(2006) 01-0002 - 07 20 1 1 2 (11 100101 ; 21 101149) : 1978 1978 2001 ; 2010 ; ; ; : ; ; 24718kg 1) 1990 26211kg 260kg 1995 2001 238kg( 1)
Elucidation of fine-scale genetic structure of sandfish (Holothuria scabra) populations in Papua New Guinea and northern Australia
 Marine and Freshwater Research, 2017, 68, 1901 1911 CSIRO 2017 Supplementary material Elucidation of fine-scale genetic structure of sandfish (Holothuria scabra) populations in Papua New Guinea and northern
Marine and Freshwater Research, 2017, 68, 1901 1911 CSIRO 2017 Supplementary material Elucidation of fine-scale genetic structure of sandfish (Holothuria scabra) populations in Papua New Guinea and northern
Project: 296 File: Title: CMC-E-600 ICD Doc No: Rev 2. Revision Date: 15 September 2010
 Project: 296 File: Title: CMC-E-600 ICD Doc No: 21029100-406 Rev 2. Revision Date: 15 September 2010 Contract No.: Revisions Table ECR/ECN LTR Description Date 0 Pre Contract draft 29 July 2010 1 Replace
Project: 296 File: Title: CMC-E-600 ICD Doc No: 21029100-406 Rev 2. Revision Date: 15 September 2010 Contract No.: Revisions Table ECR/ECN LTR Description Date 0 Pre Contract draft 29 July 2010 1 Replace
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
α 1 2- Cloning and Expression of alpha 1 2-fucosyltransferase in E. coli BIOTECHNOLOGY BULLETIN
 BIOTECHNOLOGY BULLETIN 2011 3 α 1 2-300457 PCR Helicobacter pylori HP DNA α 1 2- α 1 2-fucosyltransferase α 1 2-fuct 906 bp pet-22b + pet-fuct BL21 DE3 25 0. 1 mmol /L IPTG 4 h SDS-PAGE 33 kd α 1 2- BL21
BIOTECHNOLOGY BULLETIN 2011 3 α 1 2-300457 PCR Helicobacter pylori HP DNA α 1 2- α 1 2-fucosyltransferase α 1 2-fuct 906 bp pet-22b + pet-fuct BL21 DE3 25 0. 1 mmol /L IPTG 4 h SDS-PAGE 33 kd α 1 2- BL21
2 Composition. Invertible Mappings
 Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
stability and aromaticity in the benzonitrile H 2 O complex with Na+ or Cl
 Electronic Supplementary Data A theoretical investigation on the cooperativity effect, reduced density gradient, stability and aromaticity in the benzonitrile H 2 O complex with Na+ or Cl Jiang-Bo Xie
Electronic Supplementary Data A theoretical investigation on the cooperativity effect, reduced density gradient, stability and aromaticity in the benzonitrile H 2 O complex with Na+ or Cl Jiang-Bo Xie
CHAPTER 25 SOLVING EQUATIONS BY ITERATIVE METHODS
 CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
CHAPTER 5 SOLVING EQUATIONS BY ITERATIVE METHODS EXERCISE 104 Page 8 1. Find the positive root of the equation x + 3x 5 = 0, correct to 3 significant figures, using the method of bisection. Let f(x) =
Μέτρηση της Ρυθµικής Ικανότητας σε Μαθητές Γυµνασίου που Ασχολούνται µε Αθλητικές ραστηριότητες Συνοδευµένες ή Όχι από Μουσική
 Ερευνητική Αναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµό τόµος 3 (1), 22 28 ηµοσιεύτηκε: 11 Φεβρουαρίου 2005 Inquiries in Sport & Physical Education Volume 3 (1), 22-28 Released: February 11, 2005 www.hape.gr/emag.asp
Ερευνητική Αναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµό τόµος 3 (1), 22 28 ηµοσιεύτηκε: 11 Φεβρουαρίου 2005 Inquiries in Sport & Physical Education Volume 3 (1), 22-28 Released: February 11, 2005 www.hape.gr/emag.asp
LP N to BD* C-C = BD C-C to BD* O-H = LP* C to LP* B =5.
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2016 MS No.: CP-ART-03-2016-002134.R1 Optical Response and Gas Sequestration Properties
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2016 MS No.: CP-ART-03-2016-002134.R1 Optical Response and Gas Sequestration Properties
ΜΕΛΕΤΗ ΤΗΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΣΥΝΤΑΓΟΓΡΑΦΗΣΗΣ ΚΑΙ Η ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΕΦΑΡΜΟΓΗΣ ΤΗΣ ΣΤΗΝ ΕΛΛΑΔΑ: Ο.Α.Ε.Ε. ΠΕΡΙΦΕΡΕΙΑ ΠΕΛΟΠΟΝΝΗΣΟΥ ΚΑΣΚΑΦΕΤΟΥ ΣΩΤΗΡΙΑ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΩΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΔΙΟΙΚΗΣΗ ΤΗΣ ΥΓΕΙΑΣ ΤΕΙ ΠΕΙΡΑΙΑ ΜΕΛΕΤΗ ΤΗΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΣΥΝΤΑΓΟΓΡΑΦΗΣΗΣ ΚΑΙ Η ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΕΦΑΡΜΟΓΗΣ ΤΗΣ ΣΤΗΝ ΕΛΛΑΔΑ: Ο.Α.Ε.Ε. ΠΕΡΙΦΕΡΕΙΑ ΠΕΛΟΠΟΝΝΗΣΟΥ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΩΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΔΙΟΙΚΗΣΗ ΤΗΣ ΥΓΕΙΑΣ ΤΕΙ ΠΕΙΡΑΙΑ ΜΕΛΕΤΗ ΤΗΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΣΥΝΤΑΓΟΓΡΑΦΗΣΗΣ ΚΑΙ Η ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΕΦΑΡΜΟΓΗΣ ΤΗΣ ΣΤΗΝ ΕΛΛΑΔΑ: Ο.Α.Ε.Ε. ΠΕΡΙΦΕΡΕΙΑ ΠΕΛΟΠΟΝΝΗΣΟΥ
6.1. Dirac Equation. Hamiltonian. Dirac Eq.
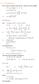 6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΤΕΧΝΟΛΟΓΙΑΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΥΠΟΛΟΓΙΣΤΩΝ Σύστημα Διαχείρισης Διαχρονικών Δεδομένων για Γονίδια ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΤΕΧΝΟΛΟΓΙΑΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΥΠΟΛΟΓΙΣΤΩΝ Σύστημα Διαχείρισης Διαχρονικών Δεδομένων για Γονίδια ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Introduction to Bioinformatics
 Introduction to Bioinformatics 260.602.01 September 2, 2005 Jonathan Pevsner, Ph.D. pevsner@jhmi.edu bioinformatics medical informatics Tool-users public health informatics databases algorithms Tool-makers
Introduction to Bioinformatics 260.602.01 September 2, 2005 Jonathan Pevsner, Ph.D. pevsner@jhmi.edu bioinformatics medical informatics Tool-users public health informatics databases algorithms Tool-makers
1 h, , CaCl 2. pelamis) 58.1%, (Headspace solid -phase microextraction and gas chromatography -mass spectrometry,hs -SPME - Vol. 15 No.
 2 0 1 5 7 Journal of Chinese Institute of Food Science and Technology Vol. 15 No. 7 Jul. 2 0 1 5 1,2 1 1 1 1* ( 1 315211 2 315502) : ( ) : (E-Nose), - : GC-MS, 20.98% 2.5%, 53.15% 11.2%, 0. 51% 71.86%
2 0 1 5 7 Journal of Chinese Institute of Food Science and Technology Vol. 15 No. 7 Jul. 2 0 1 5 1,2 1 1 1 1* ( 1 315211 2 315502) : ( ) : (E-Nose), - : GC-MS, 20.98% 2.5%, 53.15% 11.2%, 0. 51% 71.86%
Modbus basic setup notes for IO-Link AL1xxx Master Block
 n Modbus has four tables/registers where data is stored along with their associated addresses. We will be using the holding registers from address 40001 to 49999 that are R/W 16 bit/word. Two tables that
n Modbus has four tables/registers where data is stored along with their associated addresses. We will be using the holding registers from address 40001 to 49999 that are R/W 16 bit/word. Two tables that
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ :
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΕΠΙΣΤΗΜΗ & ΤΕΧΝΟΛΟΓΙΑ ΤΡΟΦΙΜΩΝ & ΔΙΑΤΡΟΦΗ ΤΟΥ ΑΝΘΡΩΠΟΥ» ΕΡΓΑΣΤΗΡΙΟ: ΧΗΜΕΙΑΣ ΚΑΙ ΑΝΑΛΥΣΗΣ ΤΡΟΦΙΜΩΝ
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΕΠΙΣΤΗΜΗ & ΤΕΧΝΟΛΟΓΙΑ ΤΡΟΦΙΜΩΝ & ΔΙΑΤΡΟΦΗ ΤΟΥ ΑΝΘΡΩΠΟΥ» ΕΡΓΑΣΤΗΡΙΟ: ΧΗΜΕΙΑΣ ΚΑΙ ΑΝΑΛΥΣΗΣ ΤΡΟΦΙΜΩΝ
Bayesian statistics. DS GA 1002 Probability and Statistics for Data Science.
 Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Θωμάς ΣΑΛΟΝΙΚΙΟΣ 1, Χρήστος ΚΑΡΑΚΩΣΤΑΣ 2, Βασίλειος ΛΕΚΙΔΗΣ 2, Μίλτων ΔΗΜΟΣΘΕΝΟΥΣ 1, Τριαντάφυλλος ΜΑΚΑΡΙΟΣ 3,
 Αξιοποίηση Έξι Σεισμών στην Πελοπόννησο για την Συσχέτιση Φασματικών Επιταχύνσεων με την Απόκριση του Δομημένου Περιβάλλοντος Correlation of Spectral Accelerations with the Response of the Built Environment
Αξιοποίηση Έξι Σεισμών στην Πελοπόννησο για την Συσχέτιση Φασματικών Επιταχύνσεων με την Απόκριση του Δομημένου Περιβάλλοντος Correlation of Spectral Accelerations with the Response of the Built Environment
SUPPORTING INFORMATION
 SUPPRTING INFRMATIN Per(-guanidino--deoxy)cyclodextrins: Synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Nikolaos Mourtzis, a Kyriaki Eliadou, a Chrysie Aggelidou,
SUPPRTING INFRMATIN Per(-guanidino--deoxy)cyclodextrins: Synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Nikolaos Mourtzis, a Kyriaki Eliadou, a Chrysie Aggelidou,
Electronic Supplementary Information DFT Characterization on the Mechanism of Water Splitting Catalyzed by Single-Ru-substituted Polyoxometalates
 Electronic Supplementary Information DFT Characterization on the Mechanism of Water Splitting Catalyzed by Single-Ru-substituted Polyoxometalates Zhong-Ling Lang, Guo-Chun Yang, Na-Na Ma, Shi-Zheng Wen,
Electronic Supplementary Information DFT Characterization on the Mechanism of Water Splitting Catalyzed by Single-Ru-substituted Polyoxometalates Zhong-Ling Lang, Guo-Chun Yang, Na-Na Ma, Shi-Zheng Wen,
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΩΡΓΙΚΗΣ ΖΩΟΛΟΓΙΑΣ ΚΑΙ ΕΝΤΟΜΟΛΟΓΙΑΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΩΡΓΙΚΗΣ ΖΩΟΛΟΓΙΑΣ ΚΑΙ ΕΝΤΟΜΟΛΟΓΙΑΣ Χρήση μοριακών δεικτών για τη διάκριση πληθυσμών των ειδών εντόμων Macrolophus pygmaeus και
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΩΡΓΙΚΗΣ ΖΩΟΛΟΓΙΑΣ ΚΑΙ ΕΝΤΟΜΟΛΟΓΙΑΣ Χρήση μοριακών δεικτών για τη διάκριση πληθυσμών των ειδών εντόμων Macrolophus pygmaeus και
Supporting information. An unusual bifunctional Tb-MOF for highly sensing of Ba 2+ ions and remarkable selectivities of CO 2 /N 2 and CO 2 /CH 4
 Electronic Supplementary Material (ESI) for Journal of Materials Chemistry A. This journal is The Royal Society of Chemistry 2015 Supporting information An unusual bifunctional Tb-MOF for highly sensing
Electronic Supplementary Material (ESI) for Journal of Materials Chemistry A. This journal is The Royal Society of Chemistry 2015 Supporting information An unusual bifunctional Tb-MOF for highly sensing
Selecting Critical Properties of Terpenes and Terpenoids through Group-Contribution Methods and Equations of State
 Supporting Information Selecting Critical Properties of Terpenes and Terpenoids through Group-Contribution Methods and Equations of State Mónia A. R. Martins, 1,2 Pedro J. Carvalho, 1 André M. Palma, 1
Supporting Information Selecting Critical Properties of Terpenes and Terpenoids through Group-Contribution Methods and Equations of State Mónia A. R. Martins, 1,2 Pedro J. Carvalho, 1 André M. Palma, 1
E#ects of Drying on Bacterial Activity and Iron Formation in Acid Sulfate Soils
 J. Jpn. Soc. Soil Phys. No. 3+, p..3 /1,**, * ** ** E#ects of Drying on Bacterial Activity and Iron Formation in Acid Sulfate Soils Kaoru UENO*, Tadashi ADACHI** and Hajime NARIOKA** * The Graduate School
J. Jpn. Soc. Soil Phys. No. 3+, p..3 /1,**, * ** ** E#ects of Drying on Bacterial Activity and Iron Formation in Acid Sulfate Soils Kaoru UENO*, Tadashi ADACHI** and Hajime NARIOKA** * The Graduate School
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor
 Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ ΨΥΧΟΛΟΓΙΚΕΣ ΕΠΙΠΤΩΣΕΙΣ ΣΕ ΓΥΝΑΙΚΕΣ ΜΕΤΑ ΑΠΟ ΜΑΣΤΕΚΤΟΜΗ ΓΕΩΡΓΙΑ ΤΡΙΣΟΚΚΑ Λευκωσία 2012 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ
ΠΕΡΙΕΧΟΜΕΝΑ. Κεφάλαιο 1: Κεφάλαιο 2: Κεφάλαιο 3:
 4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
Supporting Information. Research Center for Marine Drugs, Department of Pharmacy, State Key Laboratory
 Supporting Information Dysiherbols A C and Dysideanone E, Cytotoxic and NF-κB Inhibitory Tetracyclic Meroterpenes from a Dysidea sp. Marine Sponge Wei-Hua Jiao,, Guo-Hua Shi,, Ting-Ting Xu,, Guo-Dong Chen,
Supporting Information Dysiherbols A C and Dysideanone E, Cytotoxic and NF-κB Inhibitory Tetracyclic Meroterpenes from a Dysidea sp. Marine Sponge Wei-Hua Jiao,, Guo-Hua Shi,, Ting-Ting Xu,, Guo-Dong Chen,
Electronic Supplementary Information
 Electronic Supplementary Information Cucurbit[7]uril host-guest complexes of the histamine H 2 -receptor antagonist ranitidine Ruibing Wang and Donal H. Macartney* Department of Chemistry, Queen s University,
Electronic Supplementary Information Cucurbit[7]uril host-guest complexes of the histamine H 2 -receptor antagonist ranitidine Ruibing Wang and Donal H. Macartney* Department of Chemistry, Queen s University,
Heavier chalcogenone complexes of bismuth(iii)trihalides: Potential catalysts for acylative cleavage of cyclic ethers. Supporting Information
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2015 Heavier chalcogenone complexes of bismuth(iii)trihalides: Potential catalysts for acylative
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2015 Heavier chalcogenone complexes of bismuth(iii)trihalides: Potential catalysts for acylative
ρ. Πολιτικός Μηχανικός, ΠΑΝΓΑΙΑ ΣΥΜΒΟΥΛΟΙ ΜΗΧΑΝΙΚΟΙ Ε.Π.Ε.
 Σήραγγα ΣΓ της Ευρείας Παράκαµψης Πατρών ΣΓ Tunnel of Patras City Ring Road ΒΑΝΤΟΛΑΣ, Β. ΖΑΚΑΣ, Μ. ΡΑΧΑΝΙΩΤΗΣ, Ν. ΤΖΗΜΑ, Κ. ΤΣΑΤΣΑΝΙΦΟΣ, Χ. Πολιτικός Μηχανικός, M.Sc., ΠΑΝΓΑΙΑ ΣΥΜΒΟΥΛΟΙ ΜΗΧΑΝΙΚΟΙ Ε.Π.Ε.
Σήραγγα ΣΓ της Ευρείας Παράκαµψης Πατρών ΣΓ Tunnel of Patras City Ring Road ΒΑΝΤΟΛΑΣ, Β. ΖΑΚΑΣ, Μ. ΡΑΧΑΝΙΩΤΗΣ, Ν. ΤΖΗΜΑ, Κ. ΤΣΑΤΣΑΝΙΦΟΣ, Χ. Πολιτικός Μηχανικός, M.Sc., ΠΑΝΓΑΙΑ ΣΥΜΒΟΥΛΟΙ ΜΗΧΑΝΙΚΟΙ Ε.Π.Ε.
