Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi
|
|
|
- Διώνη Βουρδουμπάς
- 6 χρόνια πριν
- Προβολές:
Transcript
1 Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi Haiqin Chen, Guangfei Hao, Lei Wang, Hongchao Wang, Zhennan Gu, Liming Liu, Hao Zhang, Wei Chen, and Yong Q. Chen Fold change Genes Log Fold -4 : 4 A B E K L M a b A B E K L M c d e f g h Figure S Overall transcriptional regulation during lipogenesis in M. alpina. Gene transcription changes were compared among samples A, B, E, K, L and M using
2 sample E (3 min prior to nitrogen exhaustion) as the reference point. The smallest non zero FPKM was.53. Therefore,.5 FPKM was considered as the detection limit and used to replace for log transformation. The number of genes with >5 fold change is indicated at the top. Hierarchical clustering of differentially expressed genes (3328) and samples (A, B, E, K, L, and M) is shown on the left. Eight gene clusters (a, b, c, d, e, f, g, h) were identified and the average expression pattern for each cluster is shown on the right.
3 Group Gene PK (2.7..4) PCK (4...49) 4 PDH (.2.4.) 5 OGDH (.2.4.2) SUCLG (6.2..4) 7 ACLY ( ) 8 ACS (6.2..) 9 ALDH (.2..3) ADH (...2) GPAT (2.3..5) DGAT (2.3..2) ACSL (6.2..3) G6PD (...49) PGD (...44) HK (2.7.., ALDO (4..2.3) GAPDH (.2..2) ENO (4.2..) ACO (4.2..3) IDH (...42) 6 MDH (...37) 2 CRAT (2.3..7) 3 LIP (3...3) up-regulation down-regulation unchanged not identified M. alpina M. circinelloides R. oryzae C. reinhardtii A. nidulans similarity % 95% 75% 36% Figure S2 Similarity in core gene expression. Transcriptional regulation of the 32 core genes involved in carbon flux and NADPH metabolism illustrated in Figure 2 were compared among M. circinelloides, R.oryzae, C. reinhardtii and A. nidulans and M. alpina.
4 Expression level (log 2 -ΔΔCt ) Expression level (log 2 -ΔΔCt ) PK (2.7..4) 2 PCK (4...32, ) 7. ACLY ( ) Expression level (log 2 -ΔΔCt ) - 2 ACO (4.2..3) IDH (...42) - 8. ACS (6.2..) HK/GCK (2.7.., ) PDH (.2.4.) - ALDO (4..2.3) ALDH (.2..3) ADH (...2) OGDH (.2.4.2) SUCLG (6.2..4) GAPDH (.2..2) ACSL (6.2..3) CRAT (2.3..7).5 LIP (3...3) G6PD (...49) PGD (...44) A B E K L M A B E K L M A B E K L M A B E K L M A B E K L M Expression level (log 2 -ΔΔCt ) ENO (4.2..) MDH (...37) GPAT (2.3..5) DGAT (2.3..2) Figure S3 Expression similarities of the core genes in M. alpina, M. circinelloides and R. oryzae. The twenty-three genes involved in carbon flux and NADPH metabolism illustrated in Figure 2 of M. alpina, M. circinelloides and R. oryzae were analyzed by qrt-pcr during lipogenesis. Different symbols indicate different genes with the same annotation. The error bars represent standard deviations. The full names of the enzymes are indicated in the legend of Figure 2.
5 .4.2 AA (nmol/g DW /h) WT ΔG6PD ΔPGD ΔALDH4A ΔPYCR ΔNIT-6 ΔargC ΔHMGCR Figure S4 The effect of different NADPH-generating genes on AA production. The effect of different NADPH generating genes on AA production were simulated by single gene deletion using MOMA. WT: wide type, G6PD: glucose-6-phosphate dehydrogenase (EC...49), PGD: phosphogluconate dehydrogenase (EC...44), ALDH4A: -pyrroline-5-carboxylate dehydrogenase (EC.5..2), PYCR: pyrroline-5- carboxylate (EC.5..2), NIT-6: nitrite (EC.7..4), argc: N- Acetyl-gamma-glutamyl-phosphate (EC.2..38), HMGCR: hydroxymethylglutaryl-coa (EC...34).
6 G6PD G6PD2 G6PD3 PGD ME ME2 Expression level (change fold) MA-G6PD-S MA-G6PD-S2 MA-G6PD-S3 MA-G6PD-S MA-G6PD-S2 MA-G6PD-S3 MA-G6PD-S MA-G6PD-S2 MA-G6PD-S3 MA-PGD-S MA-PGD-S2 MA-PGD-S3 MA-ME-S MA-ME-S2 MA-ME-S3 MA-ME2-S MA-ME2-S2 MA-ME2-S3 Figure S5 Confirmation of RNA knockdown. Three mutants from each RNAi experiments were analyzed by qrt-pcr with the wild-type M. alpina as control. Samples were taken from cultures growing in 5 ml flasks with ml of Kendrick medium stirred at 2 rpm for 96 h at 28 C. Three independent experiments were performed and the bars represent the standard deviations. p <.5 compared to the wild-type control.
7 Table S. Summary of transcriptome data Library Sample A B E K L M Raw RNA sequencing reads (million) Mapped RNA sequencing reads (million) Predicted gene models covered A: 2 h, B: -2 h, E: -3 min, K: + h, L: +2 h and M: +48 h
8 Table S2. Predicted NADPH anabolic reactions Rxn EC Enzyme Formula μ=.3 R.2..5 aldehyde dehydrogenase [NAD(P)[c] +] + nadp[c] + acal[c] -> 2 h[c] + nadph[c] + ac[c].4 R Isocitrate dehydrogenase (NADP[c] +) nadp[c] + icit[c] -> akg[c] + nadph[c] + co2[c] R Isocitrate dehydrogenase (NADP[c] +) nadp[c] + icit[c] -> akg[c] + nadph[c] + co2[c] R glucose-6-phosphate dehydrogenase nadp[c] + dg6p[c] <=> h[c] + nadph[c] + d6pgl[c].4 R Phosphogluconate dehydrogenase nadp[c] + d6pgc[c] -> co2[c] + nadph[c] + rl5p[c].4 (decarboxylating) R6...4 malate dehydrogenase (oxaloacetatedecarboxylating) nadp[m] + mal[m] -> pyr[m] + nadph[m] + co2[m].555 (NADP[m] +) R7...4 malate dehydrogenase (oxaloacetatedecarboxylating) nadp[c] + mal[c] -> pyr[c] + nadph[c] + co2[c] (NADP[m] +) R8...- unknown hcoa[m] + nadp[m] -> nadph[m] + h[m] + 3ocoa[m] R9 no ec malcoa[m] + nadph[m] + 2 h[m] <=> 3ocoa[m] + nadp[m] + h2o[m] R no ec malcoa[c] + nadph[c] + 2 h[c] <=> 3ocoa[c] + nadp[c] + R...- unknown hcoa[m] + nadp[m] -> nadph[m] + h[m] + 3ocoa[m] R succinate-semialdehyde dehydrogenase h2o[m] + succsal[m] + nadp[m] -> 2 h[m] + nadph[m] + succ[m] R3...2 L-arabinose h[c] + nadph[c] + larab[c] <=> nadp[c] + laol[c] R4...2 D-Ribose h[c] + nadph[c] + rib[c] <=> nadp[c] + ribol[c] R pyrroline-5-carboxylate dehydrogenase sp5c[c] + nadp[c] + 2 -> glu[c] + nadph[c] + h[c] R pyrroline-5-carboxylate dehydrogenase sp5c[m] + nadp[m] + 2 h2o[m] -> glu[m] + nadph[m] + h[m] R pyrroline-5-carboxylate dehydrogenase glugsal[c] + nadp[c] + -> glu[c] + nadph[c] + 2 h[c].56 R glutamate-5-semialdehyde dehydrogenase 2 h[c] + nadph[c] + glup[c] <=> nadp[c] + pi[c] + glugsal[c] R pyrroline-5-carboxylate dehydrogenase sp5c[c] + nadp[c] + 2 -> glu[c] + nadph[c] + h[c] R Pyrroline-5-carboxylate pro[c] + nadp[c] <=> 2 h[c] + nadph[c] + sp5c[c].56 R Prephenate dehydrogenase (NADP[c] +) nadp[c] + phen[c] -> co2[c] + nadph[c] + 4hppyr[c] R aspartate dehydrogenase asp[c] + nadp[c] -> 2 h[c] + nadph[c] + iasp[c].3 R ferredoxin-nadp+ nadp[c] + h[c] + refdox[c] -> nadph[c] + oxfdox[c] R Dihydrofolate nadp[c] + dhf[c] <=> h[c] + nadph[c] + fot[c] R ; pyruvoyltetrahydropterin synthase/sepiapterin ahtd[c] + 2 nadph[c] + 2 h[c] -> bh4[c] + 2 nadp[c] + pppi[c] R ,7-dihydropteridine bh4[c] + nadp[c] -> bh2[c] + nadph[c] + h[c]
9 R methylenetetrahydrofolate dehydrogenase nadp[c] + metthf[c] <=> nadph[c] + methf[c].9 (NADP[c] +) R Oxidos ccdol[m] + nadp[m] -> techol[m] + nadph[m] + h[m] R Oxidos cdtbe[m] + nad[m] -> dtbe[m] + nadh[m] + h[m] R3...2 alcohol dehydrogenase (NADP[c] +) 6hyac[c] + nadp[c] -> adsde[c] + nadph[c] + h[c] R aldehyde dehydrogenase [NAD(P)[m] +] aldmde[m] + nadp[m] + h2o[m] -> capmde[m] + nadph[m] + h[m] R salicylate hydroxylase + co2[c] + nadp[c] + ccl[c] <=> o2[c] + 2 h[c] + nadph[c] + sali[c] R aldehyde dehydrogenase [NAD(P)[c] +] + nadp[c] + acal[c] -> 2 h[c] + nadph[c] + ac[c].4 R C-4 sterol decarboxylase (cg26);sterol-4alphacarboxylate nadp[c] + dcda[c] -> co2[c] + nadph[c] + cdol[c] 3-dehydrogenase (decarboxylating) R lathosterol oxidase epst[c] + nadp[c] -> ergod[c] + nadph[c] + h[c] R36...2;...72 NADP-dependent alcohol nadp[c] + gl[c] <=> h[c] + nadph[c] + glyal[c] dehydrogenase/glycerol dehydrogenase R Saccharopine dehydrogenase (NADP[c] +, L- glutamate forming) h2o[m] + nadp[m] + sacp[m] <=> glu[m] + h[m] + nadph[m] + amasa[m].5
10 Table S3. Predicted NADPH catabolic reactions Rxn EC Enzyme Formular μ=.3 R...2 Alcohol dehydrogenase (NADP[c] +) h[c] + nadph[c] + acal[c] -> nadp[c] + eth[c] R GDP-L-fucose synthase gdman[c] + nadph[c] + h[c] -> glfuc[c] + nadp[c] R glyoxylate (NADP[c] +) glx[c] + nadph[c] + h[c] -> glya[c] + nadp[c] R4...2 D-Xylose (xyra) (xylitol h[c] + nadph[c] + xyl[c] -> nadp[c] + xol[c] dehydrogenase) R glutamate synthase (NADH) akg[c] + gln[c] + h[c] + nadph[c] -> 2 glu[c] + nadp[c] R glutamate dehydrogenase [NAD(P)[m] +] akg[m] + nh3[m] + nadph[m] + h[m] -> glu[m] + nadp[m] + h2o[m] R glutamate dehydrogenase [NAD(P)+] akg[c] + nh3[c] + nadph[c] + h[c] -> glu[c] + nadp[c] + R (R)-2,3-Dihydroxy-3-methylbutanoate:NADP+ hmobut[m] + nadph[m] + h[m] -> nadp[m] + dmbut[m] oxido R L-aminoadipate-semialdehyde dehydrogenase 2 h[m] + nadph[m] + ama[m] -> h2o[m] + nadp[m] + amasa[m] R.5..2 Pyrroline-5-carboxylate 2 h[c] + nadph[c] + phc[c] -> hpro[c] + nadp[c] R Kynurenine 3-monooxygenase o2[c] + h[c] + nadph[c] + kyn[c] -> + nadp[c] + hkyn[c] R shikimate dehydrogenase h[c] + nadph[c] + dhsk[c] -> nadp[c] + sme[c] R (R)-2,3-Dihydroxy-3-methylpentanoate:NADP+ rhmopt[m] + nadph[m] + h[m] -> dmvat[m] + nadp[m].6 oxido (isomerizing) R (R)-2,3-Dihydroxy-3-methylpentanoate:NADP+ rhmopt[c] + nadph[c] + h[c] -> dmvat[c] + nadp[c] oxido (isomerizing) R thioredoxin (NADPH) h[c] + nadph[c] + othio[c] -> nadp[c] + rthio[c] R glutathione-disulfide h[c] + nadph[c] + gssg[c] -> nadp[c] + 2 gsh[c] R nitrite [NADPH] 5 h[c] + 3 nadph[c] + hno2[c] -> + 3 nadp[c] + nh4oh[c] R Sulfite (NADPH) 3 h[c] + 3 nadph[c] + slfi[c] -> nadp[c] + h2s[c] R pyridoxine 4-dehydrogenase pdxal[c] + nadph[c] + h[c] -> vb6[c] + nadp[c] R dehydropantoate 2- h[m] + nadph[m] + akp[m] -> nadp[m] + pant[m].3 R2... B-ketoacyl-ACP synthase (c,), fatty acyl 6 h[c] + 4 nadph[c] + 3 malcoa[c] -> co2[c] + 4 CoA synthase R22... B-ketoacyl-ACP synthase (c,), fatty acyl CoA synthase nadp[c] + 2 coa[c] + chcoa[c] 6 h[m] + 4 nadph[m] + 3 malcoa[m] -> 2 h2o[m] + 2 co2[m] + 4 nadp[m] + 2 coa[m] + chcoa[m]
11 R Dihydrofolate h[c] + nadph[c] + dhf[c] -> thf[c] + nadp[c] R Dihydrofolate 2 h[c] + 2 nadph[c] + fot[c] -> 2 nadp[c] + thf[c] R octaprenyl-6-methoxyphenol hydroxylase o2[m] + nadph[m] + hm[m] -> h2o[m] + nadp[m] + h[m] + hmb[m] R ubiquinone biosynthesis monooxygenase hmmb[m] + o2[m] + nadph[m] + h[m] -> hmhmb[m] + Coq7 nadp[m] + h2o[m] R Squalene synthase 2 fpp[c] + nadph[c] + 3 h[c] -> 2 ppi[c] + sql[c] + nadp[c] R squalene monooxygenase sql[c] + o2[c] + nadph[c] + h[c] -> s23e[c] + nadp[c] + R zeaxanthin epoxidase atxin[c] + nadph[c] + h[c] + o2[c] -> vlxin[c] + nadp[c] + R zeaxanthin epoxidase zextin[c] + nadph[c] + h[c] + o2[c] -> axtin[c] + nadp[c] + R dtdp-4-dehydrorhamnose dtdp6m[c] + h[c] + nadph[c] -> nadp[c] + dtdpdm[c] R hydroxybutyryl-CoA dehydrogenase c4hcoa[c] + nadp[c] <=> aaccoa[c] + nadph[c] + h[c] R Sterigmatocystin biosynthesis monooxygenase o2[m] + h[m] + nadph[m] + avn[m] -> h2o[m] + nadp[m] + havn[m] R Sterigmatocystin biosynthesis monooxygenase o2[c] + h[c] + nadph[c] + avn[c] -> + nadp[c] + havn[c] R phenol 2-monooxygenase 3csol[m] + o2[m] + nadph[m] + h[m] -> 23dlne[m] + nadp[m] + h2o[m] R phenol 2-monooxygenase pnol[m] + o2[m] + nadph[m] + h[m] -> ccl[m] + nadp[m] + h2o[m] R phenol 2-monooxygenase rsnol[m] + o2[m] + nadph[m] + h[m] -> btol[m] + nadp[m] + h2o[m] R phenol 2-monooxygenase 3csol[c] + o2[c] + nadph[c] + h[c] -> 23dlne[c] + nadp[c] + R phenol 2-monooxygenase pnol[c] + o2[c] + nadph[c] + h[c] -> ccl[c] + nadp[c] + R phenol 2-monooxygenase rsnol[c] + o2[c] + nadph[c] + h[c] -> btol[c] + nadp[c] + R Oxidos 4eol[m] + nadph[m] + h[m] + o2[m] -> eal[m] + nadp[m] + h2o[m] R Oxidos bendl[m] + nadph[m] + h[m] + o2[m] -> btol[m] + nadp[m] + h2o[m] R Oxidos sali[m] + nadph[m] + o2[m] + h[m] -> 25dhba[m] + nadp[m] + h2o[m] R dimethylaniline monooxygenase tafen[c] + o2[c] + nadph[c] + h[c] -> tafnox[c] + nadp[c] +
12 R dimethylaniline monooxygenase tafen[p] + o2[p] + nadph[p] + h[p] -> tafnox[p] + nadp[p] + h2o[p] R Aflatoxins biosynthesis;,3,6,8-tetra hydroxy h[c] + nadph[c] + vera[c] -> nadp[c] + dmst[c] naphthalene R Aflatoxins biosynthesis;,3,6,8-tetra hydroxy h[c] + nadph[c] + verb[c] -> + nadp[c] + dhdmst[c] naphthalene R48... beta-ketoacyl-[acyl-carrier protein](acp) c4hacp[m] + nadp[m] <=> aacacp[m] + nadph[m] + h[m] R49... beta-ketoacyl-[acyl-carrier protein](acp) c6hacp[m] + nadp[m] <=> c6oacp[m] + nadph[m] + h[m] R5... beta-ketoacyl-[acyl-carrier protein](acp) c8hacp[m] + nadp[m] <=> c8oacp[m] + nadph[m] + h[m] R5... beta-ketoacyl-[acyl-carrier protein](acp) chacp[m] + nadp[m] <=> coacp[m] + nadph[m] + h[m] R52... beta-ketoacyl-[acyl-carrier protein](acp) c2hacp[m] + nadp[m] <=> c2oacp[m] + nadph[m] + h[m] R53... beta-ketoacyl-[acyl-carrier protein](acp) c4hacp[m] + nadp[m] <=> c4oacp[m] + nadph[m] + h[m] R54... beta-ketoacyl-[acyl-carrier protein](acp) c6hacp[m] + nadp[m] <=> c6oacp[m] + nadph[m] + h[m] R oxoacyl-[acyl-carrier-protein] aacacp[c] + nadph[c] + h[c] <=> c4hacp[c] + nadp[c].62 R fatty-acyl-coa synthase c4dacp[c] + nadph[c] + h[c] <=> c4acp[c] + nadp[c].62 R oxoacyl-[acyl-carrier-protein] c6oacp[c] + nadph[c] + h[c] <=> c6hacp[c] + nadp[c].62 R fatty-acyl-coa synthase c6dacp[c] + nadph[c] + h[c] <=> c6acp[c] + nadp[c].62 R oxoacyl-[acyl-carrier-protein] c8oacp[c] + nadph[c] + h[c] <=> c8hacp[c] + nadp[c].62 R fatty-acyl-coa synthase c8dacp[c] + nadph[c] + h[c] <=> c8acp[c] + nadp[c].62 R oxoacyl-[acyl-carrier-protein] coacp[c] + nadph[c] + h[c] <=> chacp[c] + nadp[c].62 R fatty-acyl-coa synthase cdacp[c] + nadph[c] + h[c] <=> cacp[c] + nadp[c].62 R oxoacyl-[acyl-carrier-protein] c2oacp[c] + nadph[c] + h[c] <=> c2hacp[c] + nadp[c].62 R fatty-acyl-coa synthase c2dacp[c] + nadph[c] + h[c] <=> c2acp[c] + nadp[c].62 R oxoacyl-[acyl-carrier-protein] c4oacp[c] + nadph[c] + h[c] <=> c4hacp[c] + nadp[c].62 R fatty-acyl-coa synthase c4dacp[c] + nadph[c] + h[c] <=> c4acp[c] + nadp[c].62 R oxoacyl-[acyl-carrier-protein] c6oacp[c] + nadph[c] + h[c] <=> c6hacp[c] + nadp[c].62 R fatty-acyl-coa synthase c6dacp[c] + nadph[c] + h[c] <=> c6acp[c] + nadp[c].62
13 R very-long-chain 3-oxoacyl-CoA c8ocoa[c] + nadph[c] + h[c] <=> c8hcoa[c] + nadp[c].56 R very-long-chain enoyl-coa c8dcoa[c] + nadph[c] + h[c] <=> c8coa[c] + nadp[c].56 R very-long-chain 3-oxoacyl-CoA c2ocoa[c] + nadph[c] + h[c] <=> c2hcoa[c] + nadp[c].2 R very-long-chain enoyl-coa c2dcoa[c] + nadph[c] + h[c] <=> c2coa[c] + nadp[c].2 R very-long-chain 3-oxoacyl-CoA c22ocoa[c] + nadph[c] + h[c] <=> c22hcoa[c] + nadp[c]. R very-long-chain enoyl-coa c22dcoa[c] + nadph[c] + h[c] <=> c22coa[c] + nadp[c]. R very-long-chain 3-oxoacyl-CoA c24ocoa[c] + nadph[c] + h[c] <=> c24hcoa[c] + nadp[c] R very-long-chain enoyl-coa c24dcoa[c] + nadph[c] + h[c] <=> c24coa[c] + nadp[c] R sterol 4-demethylase lnst[c] + 3 o2[c] + 3 nadph[c] + 2 h[c] -> dctol[c] + for[c] + 3 nadp[c] + 4 R lanosterol delta24- lnst[c] + nadph[c] + h[c] -> dhstro[c] + nadp[c] R delta4-sterol ;C-4 sterol h[c] + nadph[c] + dctol[c] -> nadp[c] + dcdol[c] (cg24) R methylsterol monooxygenase;c-4 sterol methyl 3 o2[c] + 2 h[c] + 3 nadph[c] + dcdol[c] -> oxidase (cg25) nadp[c] + dcda[c] R keto-steroid h[c] + nadph[c] + cdol[c] -> nadp[c] + mzymst[c] R delta24-sterol zymst[c] + nadph[c] + h[c] -> ac8bol[c] + nadp[c] R lathosterol oxidase ac7bol[c] + nadph[c] + h[c] + o2[c] -> pvd3[c] + nadp[c] + 2 R delta24-sterol acdbol[c] + nadph[c] + h[c] -> ac7bol[c] + nadp R dehydrocholesterol pvd3[c] + nadph[c] + h[c] -> chtrol[c] + nadp[c] R delta24-sterol 7strol[c] + nadph[c] + h[c] -> pvd3[c] + nadp[c] R sterol 4-demethylase obl[c] + 3 o2[c] + 3 nadph[c] + 2 h[c] -> amt3bol[c] + for[c] + 3 nadp[c] + 4 R Delta4-sterol amt3bol[c] + nadph[c] + h[c] -> mefol[c] + nadp[c] R dehydrocholesterol dhrsol[c] + nadph[c] + h[c] -> isol[c] + nadp[c] R delta24(24())-sterol h[c] + nadph[c] + egteol[c] -> nadp[c] + egstr[c] R C-5 sterol desaturase (ERG3) nadph[c] + epst[c] -> 3 h[c] + nadp[c] + ergod[c] R lathosterol oxidase acdbol[c] + nadph[c] -> 7dstrol[c] + nadp[c] + 3 h[c] R dehydrocholesterol 7dstrol[c] + nadph[c] + h[c] -> nadp[c] + dmstrol[c] R94... Acylglycerone-phosphate 4 h[c] + nadph[c] + at3p2[c] -> nadp[c] + agl3p[c] R Dehydrosphinganine h[c] + nadph[c] + dhsph[c] -> nadp[c] + sph[c] R C4-hydroxylase sph[c] + nadph[c] + h[c] + o2[c] -> nadp[c] + psph[c] +
14 R C4-hydroxylase o2[c] + h[c] + nadph[c] + dcer2[c] -> + nadp[c] + pcer2[c] R hexadecanal:nadp+ delta2-oxido c6e[c] + nadph[c] + h[c] -> c6a[c] + nadp[c] R sphingolipid delta-4 desaturase o2[c] + h[c] + nadph[c] + dcer2[c] -> + nadp[c] + pcer2[c] R.4.4. dimethylallyl diphosphate c24(6)[c] + o2 + nadph[c] + 2 h[c] -> 45eet[c] + nadp[c] + R.4.4. Cytochrome P45 c24(6)[c] + o2 + nadph[c] + 2 h[c] -> 2eet[c] + nadp[c] + R Cytochrome P45 c24(6)[c] + o2 + nadph[c] + 2 h[c] -> 89eet[c] + nadp[c] + R Cytochrome P45 c24(6)[c] + o2 + nadph[c] + 2 h[c] -> 56eet[c] + nadp[c] + R Cytochrome P45 c24(6)[c] + o2 + nadph[c] + 2 h[c] -> 6hete[c] + nadp[c] + R delta 9 desaturase c8acp[m] + o2[m] + nadph[m] + 3 h[m] -> c8acp[m] + 2 h2o[m] + nadp[m] R NADPH--cytochrome P45 nadph[c] + 2 feri[m] -> h[m] + nadp[c] + 2 fero[m] R7.4.- C4-hydroxylase sph[c] + nadph[c] + h[c] + o2[c] -> nadp[c] + psph[c] + R delta 9 desaturase c6coa[c] + o2[c] + nadph[c] + h[c] -> c6coa[c] nadp[c] R delta 9 desaturase c8coa[c] + o2[c] + nadph[c] + h[c] -> c8coa[c] nadp[c] R delta 2 desaturase c8coa[c] + o2[c] + nadph[c] + h[c] -> c82coa[c] nadp[c] R delta 5 desaturase c82coa[c] + o2[c] + nadph[c] + h[c] -> c83(3)coa[c] nadp[c] R delta 6 desaturase c82coa[c] + o2[c] + nadph[c] + h[c] -> c83(6)coa[c] +.39 nadp[c] + 2 R delta 6 desaturase c83(3)coa[c] + o2[c] + nadph[c] + h[c] -> c84(3)coa[c] + nadp[c] + 2 R delta 9 desaturase c2coa[c] + o2[c] + nadph[c] + h[c] -> c2coa[c] nadp[c] R very-long-chain 3-oxoacyl-CoA c22ocoa[c] + nadph[c] + h[c] <=> c22hcoa[c] + nadp[c]
15 R very-long-chain enoyl-coa c22dcoa[c] + nadph[c] + h[c] <=> c22coa[c] + nadp[c] R very-long-chain 3-oxoacyl-CoA c23(6)ocoa[c] + nadph[c] + h[c] <=> c23(6)hcoa[c] +.35 nadp[c] R very-long-chain enoyl-coa c23(6)dcoa[c] + nadph[c] + h[c] <=> c23(6)coa[c] +.35 nadp[c] R very-long-chain 3-oxoacyl-CoA c24(3)ocoa[c] + nadph[c] + h[c] <=> c24(3)hcoa[c] + nadp[c] R very-long-chain enoyl-coa c24(3)hcoa[c] <=> c24(3)dcoa[c] + R delta 5 desaturase c23(6)coa[c] + o2[c] + nadph[c] + h[c] -> c24(6)coa[c] +.35 nadp[c] + 2 R delta 5 desaturase c24(3)coa[c] + 2 o2[c] + nadph[c] + 3 h[c] -> c25(3)coa[c] + nadp[c] + 4 R delta 5 desaturase c24(6)coa[c] + o2[c] + nadph[c] + h[c] -> c25(3)coa[c] nadp[c] R very-long-chain 3-oxoacyl-CoA c225(3)ocoa[c] + nadph[c] + h[c] <=> c225(3)hcoa[c] + nadp[c] R very-long-chain enoyl-coa c225(3)hcoa[c] <=> c225(3)dcoa[c] + R very-long-chain 3-oxoacyl-CoA c245(3)ocoa[c] + nadph[c] + h[c] <=> c245(3)hcoa[c] + nadp[c] R very-long-chain enoyl-coa c245(3)hcoa[c] <=> c245(3)dcoa[c] + R delta 6 desaturase c245(3)coa[c] + o2[c] + nadph[c] + h[c] -> c246(3)coa[c] + nadp[c] + 2 R N-Acetyl-gamma-glutamyl-phosphate 2 h[m] + nadph[m] + naglup[m] <=> nadp[m] + naglus[m] pi[m] R3.2.. aspartate-semialdehyde dehydrogenase 2 h[c] + nadph[c] + basp[c] <=> nadp[c] + pi[c] + aspsa[c] R3...3 homoserine dehydrogenase h[m] + nadph[m] + aspsa[m] <=> nadp[m] + hser[m] R homoserine dehydrogenase h[c] + nadph[c] + aspsa[c] <=> nadp[c] + hser[c] R malonate-semialdehyde dehydrogenase oppa[m] + nadp[m] + coa[m] <=> h[m] + nadph[m] + malcoa[m] R methylenetetrahydrofolate h[c] + nadph[c] + metthf[c] <=> nadp[c] + mthf[c] [NAD(P)H] R hydroxymethylglutaryl-coa (NADPH) 2 h[c] + 2 nadph[c] + hmgcoa[c] <=> 2 nadp[c] + coa[c] + mvl[c].62
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver.
 Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Supporting Information. Experimental section
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
ΙΑΤΑΡΑΧΕΣ ΜΕΤΑΒΟΛΙΣΜΟΥ ΛΙΠΑΡΩΝ ΟΞΕΩΝ
 Ηµ/νία Εξέτασης : ΙΑΤΑΡΑΧΕΣ ΜΕΤΑΒΟΛΙΣΜΟΥ ΛΙΠΑΡΩΝ ΟΞΕΩΝ Medium Chain Acyl CoA Dehydrogenase C6< 0.24 C8< 0.35 C10< 0.3 C10:1 < 0.22 C8/C2< 0.02 C8/C10 < 3 Long Chain 3 Hydroxyacyl Coa Dehydrogenase C16OH
Ηµ/νία Εξέτασης : ΙΑΤΑΡΑΧΕΣ ΜΕΤΑΒΟΛΙΣΜΟΥ ΛΙΠΑΡΩΝ ΟΞΕΩΝ Medium Chain Acyl CoA Dehydrogenase C6< 0.24 C8< 0.35 C10< 0.3 C10:1 < 0.22 C8/C2< 0.02 C8/C10 < 3 Long Chain 3 Hydroxyacyl Coa Dehydrogenase C16OH
Selecting Critical Properties of Terpenes and Terpenoids through Group-Contribution Methods and Equations of State
 Supporting Information Selecting Critical Properties of Terpenes and Terpenoids through Group-Contribution Methods and Equations of State Mónia A. R. Martins, 1,2 Pedro J. Carvalho, 1 André M. Palma, 1
Supporting Information Selecting Critical Properties of Terpenes and Terpenoids through Group-Contribution Methods and Equations of State Mónia A. R. Martins, 1,2 Pedro J. Carvalho, 1 André M. Palma, 1
Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions
 This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
This journal is The Royal Society of Chemistry 213 Synthesis of Imines from Amines in Aliphatic Alcohols on Pd/ZrO 2 Catalyst at Ambient Conditions Wenjing Cui, a Bao Zhaorigetu,* a Meilin Jia, a and Wulan
Sar1B-mediated Regulation of Triglyceride and Cholesterol
 FIGURE S1. Comparing Sar1A- and Sar1B-flag Over-expression on RNA Profiles of McArdle RH7777 cells. RNA was prepared from two Sar1B:H79G-flag cell lines and two control cell lines plus two Sar1B-flag and
FIGURE S1. Comparing Sar1A- and Sar1B-flag Over-expression on RNA Profiles of McArdle RH7777 cells. RNA was prepared from two Sar1B:H79G-flag cell lines and two control cell lines plus two Sar1B-flag and
Guo_Fig. S1. Atg7 +/+ Atg7 -/- FBP_M+6 DHAP_M+3. Glucose Uptake Rate G6P_M+6. nmol/ul cell/hr
 Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
J. Dairy Sci. 93: doi: /jds American Dairy Science Association, 2010.
 Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
Mean bond enthalpy Standard enthalpy of formation Bond N H N N N N H O O O
 Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
Q1. (a) Explain the meaning of the terms mean bond enthalpy and standard enthalpy of formation. Mean bond enthalpy... Standard enthalpy of formation... (5) (b) Some mean bond enthalpies are given below.
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών
 Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Table S1: Inpatient Diet Composition
 Table S1: Inpatient Diet Composition Diet Components (% of Energy) Inpatient Period Baseline Intervention Protein 15.1 ± 0.0 15.0 ± 0.1 Total fat 30.0 ± 0.0 30.0 ± 0.1 Saturated fat 8.6 ± 0.8 8.4 ± 0.9
Table S1: Inpatient Diet Composition Diet Components (% of Energy) Inpatient Period Baseline Intervention Protein 15.1 ± 0.0 15.0 ± 0.1 Total fat 30.0 ± 0.0 30.0 ± 0.1 Saturated fat 8.6 ± 0.8 8.4 ± 0.9
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Identification of Fish Species using DNA Method
 5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
5 * * * Identification of Fish Species using DNA Method Yuumi KAWASHIMA*, Takayuki KATAYAMA* and Yukihiko YAMAZAKI* *Central Customs Laboratory, Ministry of Finance 6-3-5, Kashiwanoha, Kashiwa, Chiba 277-0882,
abs acces acces Sample test results. Actual results may vary. Antioxidants B-Vitamins Minerals Order Today At Results Overview
 Order Today At www.accesalabs. Antioxidants B-Vitamins Minerals Vitamin C Pyridoxine - Biotin Vitamin A / Carotenoids Vitamin E / Tocopher α-lipoic A Co T ami B1 cer R bo a n - B2 N acin - B3 Results Overview
Order Today At www.accesalabs. Antioxidants B-Vitamins Minerals Vitamin C Pyridoxine - Biotin Vitamin A / Carotenoids Vitamin E / Tocopher α-lipoic A Co T ami B1 cer R bo a n - B2 N acin - B3 Results Overview
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Daewoo Technopark A-403, Dodang-dong, Wonmi-gu, Bucheon-city, Gyeonggido, Korea LM-80 Test Report
 LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
Figure 3 Three observations (Vp, Vs and density isosurfaces) intersecting in the PLF space. Solutions exist at the two indicated points.
 φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
Lewis Acid Catalyzed Propargylation of Arenes with O-Propargyl Trichloroacetimidate: Synthesis of 1,3-Diarylpropynes
 Supporting Information for Lewis Acid Catalyzed Propargylation of Arenes with O-Propargyl Trichloroacetimidate: Synthesis of 1,3-Diarylpropynes Changkun Li and Jianbo Wang* Beijing National Laboratory
Supporting Information for Lewis Acid Catalyzed Propargylation of Arenes with O-Propargyl Trichloroacetimidate: Synthesis of 1,3-Diarylpropynes Changkun Li and Jianbo Wang* Beijing National Laboratory
ΠΑΥΛΙΝΑ 609315 ΠΕ11 25,5 ΚΑΒΑΛΑΣ ΑΝΑΤ. ΑΤΤΙΚΗ
 ΕΛΛΕΙΜΑΤΙΚΕΣ - ΠΛΕΟΝΑΣΜΑΤΙΚΕΣ 1 1 ΑΒΑΝΙΔΗ ΑΝΝΑ 593587 ΠΕ70 14 ΚΟΡΙΝΘΙΑ Α ΑΘΗΝΩΝ 2 ΑΒΕΡΚΙΑΔΟΥ ΠΑΤΑΡΙΝΣΚΑ ΠΑΥΛΙΝΑ 609315 ΠΕ11 25,5 ΚΑΒΑΛΑΣ ΑΝΑΤ. ΑΤΤΙΚΗ 3 ΑΒΟΥΡΗ ΑΙΚΑΤΕΡΙΝΗ 590405 ΠΕ16 36,917 ΖΑΚΥΝΘΟΣ ΣΕΡΡΕΣ
ΕΛΛΕΙΜΑΤΙΚΕΣ - ΠΛΕΟΝΑΣΜΑΤΙΚΕΣ 1 1 ΑΒΑΝΙΔΗ ΑΝΝΑ 593587 ΠΕ70 14 ΚΟΡΙΝΘΙΑ Α ΑΘΗΝΩΝ 2 ΑΒΕΡΚΙΑΔΟΥ ΠΑΤΑΡΙΝΣΚΑ ΠΑΥΛΙΝΑ 609315 ΠΕ11 25,5 ΚΑΒΑΛΑΣ ΑΝΑΤ. ΑΤΤΙΚΗ 3 ΑΒΟΥΡΗ ΑΙΚΑΤΕΡΙΝΗ 590405 ΠΕ16 36,917 ΖΑΚΥΝΘΟΣ ΣΕΡΡΕΣ
Φύλλο1. ΠΕΡΙΟΧΗ ΠΡΟΣΛΗΨΗΣ ΑΒΡΑΜΙΔΟΥ ΜΑΡΙΚΑ ΔΗΜΗΤΡΙΟΣ Γ Αθηνών ΑΒΡΑΜΙΔΟΥ ΣΟΦΙΑ ΔΗΜΗΤΡΙΟΣ Λασίθι ΑΓΓΕΛΗ ΑΝΔΡΟΜΑΧΗ ΒΑΣΙΛΕΙΟΣ
 ΕΠΩΝΥΜΟ ΟΝΟΜΑ ΠΑΤΡΩΝΥΜΟ ΠΕΡΙΟΧΗ ΠΡΟΣΛΗΨΗΣ ΑΒΡΑΜΙΔΟΥ ΜΑΡΙΚΑ ΔΗΜΗΤΡΙΟΣ Γ Αθηνών ΑΒΡΑΜΙΔΟΥ ΣΟΦΙΑ ΔΗΜΗΤΡΙΟΣ Λασίθι ΑΓΓΕΛΗ ΑΝΔΡΟΜΑΧΗ ΒΑΣΙΛΕΙΟΣ Α Ανατ. Αττικής ΑΓΓΕΛΟΠΟΥΛΟΥ ΚΩΝΣΤΑΝΤΙΝΑ ΠΑΝΑΓΙΩΤΗΣ Αχαία ΑΓΓΕΛΟΠΟΥΛΟΥ
ΕΠΩΝΥΜΟ ΟΝΟΜΑ ΠΑΤΡΩΝΥΜΟ ΠΕΡΙΟΧΗ ΠΡΟΣΛΗΨΗΣ ΑΒΡΑΜΙΔΟΥ ΜΑΡΙΚΑ ΔΗΜΗΤΡΙΟΣ Γ Αθηνών ΑΒΡΑΜΙΔΟΥ ΣΟΦΙΑ ΔΗΜΗΤΡΙΟΣ Λασίθι ΑΓΓΕΛΗ ΑΝΔΡΟΜΑΧΗ ΒΑΣΙΛΕΙΟΣ Α Ανατ. Αττικής ΑΓΓΕΛΟΠΟΥΛΟΥ ΚΩΝΣΤΑΝΤΙΝΑ ΠΑΝΑΓΙΩΤΗΣ Αχαία ΑΓΓΕΛΟΠΟΥΛΟΥ
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) CPh 3 as a functional group in P-heterocyclic
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) CPh 3 as a functional group in P-heterocyclic
Science of Sericulture
 Science of Sericulture 2012 38 1 0065-0069 ISSN 0257-4799 CN 32-1115 /S E-mail CYKE@ chinajournal. net. cn 215123 25 0 ~ 24 h GSH GSSG GSH + 2GSSG GSH /GSSG S- GST TPX 23% GR 57% GSH TPX GST GSSG 61% GSH
Science of Sericulture 2012 38 1 0065-0069 ISSN 0257-4799 CN 32-1115 /S E-mail CYKE@ chinajournal. net. cn 215123 25 0 ~ 24 h GSH GSSG GSH + 2GSSG GSH /GSSG S- GST TPX 23% GR 57% GSH TPX GST GSSG 61% GSH
Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance
 POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE ΑΠΟ ΑΤΟΜΑ ΜΕ ΤΥΦΛΩΣΗ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
Table S1. All regions of WCC enrichment at p<0.001 and z score >3.09
 Table S. All regions of WCC enrichment at p3.9 N. crassa contig start nt # of reads z score promoter coding region intergenic chrvii_contig7. 776 738 5.66 frq chrvi_contig7. 696.93 vvd
Table S. All regions of WCC enrichment at p3.9 N. crassa contig start nt # of reads z score promoter coding region intergenic chrvii_contig7. 776 738 5.66 frq chrvi_contig7. 696.93 vvd
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Dietary success of a new key fish in an overfished ecosystem: evidence from fatty acid and stable isotope signatures
 The following supplement accompanies the article Dietary success of a new key fish in an overfished ecosystem: evidence from fatty acid and stable isotope signatures M. G. van der Bank 1, A. C. Utne-Palm
The following supplement accompanies the article Dietary success of a new key fish in an overfished ecosystem: evidence from fatty acid and stable isotope signatures M. G. van der Bank 1, A. C. Utne-Palm
ΠΕΡΙΦΕΡΕΙΑΚΗ ΕΝΟΤΗΤΑ ΑΤΤΙΚΗΣ ΚΕΝΤΡΙΚΟΥ ΤΟΜΕΑ 17 ΠΕ ΑΤΤΙΚΗΣ ΚΕΝΤΡΙΚΟΥ ΤΟΜΕΑ 33 ΔΕ ΑΤΤΙΚΗΣ ΚΕΝΤΡΙΚΟΥ ΤΟΜΕΑ 41 ΠΕ/ΤΕ ΑΤΤΙΚΗΣ ΚΕΝΤΡΙΚΟΥ ΤΟΜΕΑ 69 ΥΕ
 A/A 1 2 3 4 ΕΠΙΒΛΕΠΩΝ ΦΟΡΕΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΥΠΗΡΕΣΙΑ ΤΟΠΟΘΕΤΗΣΗΣ ΔΙΟΙΚΗΤΙΚΗ ΠΕΡΙΦΕΡΕΙΑ ΠΕΡΙΦΕΡΕΙΑΚΗ ΕΝΟΤΗΤΑ ΑΡΙΘΜΟΣ ΘΕΣΕΩΝ ΕΠΙΠΕΔΟ ΕΚΠΑΙΔΕΥΣΗΣ
A/A 1 2 3 4 ΕΠΙΒΛΕΠΩΝ ΦΟΡΕΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΕΚΠΑΙΔΕΥΣΗΣ Α ΑΘΗΝΑΣ ΥΠΗΡΕΣΙΑ ΤΟΠΟΘΕΤΗΣΗΣ ΔΙΟΙΚΗΤΙΚΗ ΠΕΡΙΦΕΡΕΙΑ ΠΕΡΙΦΕΡΕΙΑΚΗ ΕΝΟΤΗΤΑ ΑΡΙΘΜΟΣ ΘΕΣΕΩΝ ΕΠΙΠΕΔΟ ΕΚΠΑΙΔΕΥΣΗΣ
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Τεχνικές παρασκευής ζεόλιθου ZSM-5 από τέφρα φλοιού ρυζιού με χρήση φούρνου μικροκυμάτων και τεχνικής sol-gel
 Τεχνικές παρασκευής ζεόλιθου ZSM-5 από τέφρα φλοιού ρυζιού με χρήση φούρνου μικροκυμάτων και τεχνικής sol-gel Δέσποινα Στεφοπούλου Επιβλέπων: Κωνσταντίνος Κορδάτος Στην παρούσα διπλωματική εργασία παρασκευάστηκαν
Τεχνικές παρασκευής ζεόλιθου ZSM-5 από τέφρα φλοιού ρυζιού με χρήση φούρνου μικροκυμάτων και τεχνικής sol-gel Δέσποινα Στεφοπούλου Επιβλέπων: Κωνσταντίνος Κορδάτος Στην παρούσα διπλωματική εργασία παρασκευάστηκαν
Statistics 104: Quantitative Methods for Economics Formula and Theorem Review
 Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Table 2 Suggested prediction methods to use for a given set of available input parameters per each examined soil hydraulic property) a.
 Table 2 Suggested prediction methods to use for a given set of available input parameters per each examined soil hydraulic property) a. Reference number of model indicated in Table 1 Predicted soil hydraulic
Table 2 Suggested prediction methods to use for a given set of available input parameters per each examined soil hydraulic property) a. Reference number of model indicated in Table 1 Predicted soil hydraulic
 1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
Optimizing Microwave-assisted Extraction Process for Paprika Red Pigments Using Response Surface Methodology
 2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
PHOS π 0 analysis, for production, R AA, and Flow analysis, LHC11h
 PHOS π, ask PHOS π analysis, for production, R AA, and Flow analysis, Henrik Qvigstad henrik.qvigstad@fys.uio.no University of Oslo --5 PHOS π, ask ask he task we use, AliaskPiFlow was written prior, for
PHOS π, ask PHOS π analysis, for production, R AA, and Flow analysis, Henrik Qvigstad henrik.qvigstad@fys.uio.no University of Oslo --5 PHOS π, ask ask he task we use, AliaskPiFlow was written prior, for
Supporting Information
 Supporting Information Molecular Cage Impregnated Palladium Nanoparticles: Efficient, Additive-Free Heterogeneous Catalysts for Cyanation of Aryl Halides. Bijnaneswar Mondal, Koushik Acharyya, Prodip Howlader
Supporting Information Molecular Cage Impregnated Palladium Nanoparticles: Efficient, Additive-Free Heterogeneous Catalysts for Cyanation of Aryl Halides. Bijnaneswar Mondal, Koushik Acharyya, Prodip Howlader
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supporting Information
 Tissue Level Diet and Sex-by-Diet Interactions Reveal Unique Metabolite and Clustering Profiles Using Untargeted Liquid Chromatography-Mass Spectrometry on Adipose, Skeletal Muscle, and Liver Tissue in
Tissue Level Diet and Sex-by-Diet Interactions Reveal Unique Metabolite and Clustering Profiles Using Untargeted Liquid Chromatography-Mass Spectrometry on Adipose, Skeletal Muscle, and Liver Tissue in
Supporting Information One-Pot Approach to Chiral Chromenes via Enantioselective Organocatalytic Domino Oxa-Michael-Aldol Reaction
 Supporting Information ne-pot Approach to Chiral Chromenes via Enantioselective rganocatalytic Domino xa-michael-aldol Reaction Hao Li, Jian Wang, Timiyin E-Nunu, Liansuo Zu, Wei Jiang, Shaohua Wei, *
Supporting Information ne-pot Approach to Chiral Chromenes via Enantioselective rganocatalytic Domino xa-michael-aldol Reaction Hao Li, Jian Wang, Timiyin E-Nunu, Liansuo Zu, Wei Jiang, Shaohua Wei, *
Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)]
![Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)] Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)]](/thumbs/89/98928029.jpg) d Suva refrigerants Technical Information T-9100SI Thermodynamic Properties of Suva 9100 Refrigerant [R-410A (50/50)] Thermodynamic Properties of Suva 9100 Refrigerant SI Units New tables of the thermodynamic
d Suva refrigerants Technical Information T-9100SI Thermodynamic Properties of Suva 9100 Refrigerant [R-410A (50/50)] Thermodynamic Properties of Suva 9100 Refrigerant SI Units New tables of the thermodynamic
Supplementary Materials: Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TOF-MS n
 S1 of S26 Supplementary Materials: Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TF-MS n Ping Yang, Feng Xu, Hong-Fu Li, Yi-Wang, Feng-Chun Li, Ming-Ying Shang,
S1 of S26 Supplementary Materials: Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TF-MS n Ping Yang, Feng Xu, Hong-Fu Li, Yi-Wang, Feng-Chun Li, Ming-Ying Shang,
ΑΠΟΜΟΝΩΣΗ, ΤΑΥΤΟΠΟΙΗΣΗ ΜΕΘΑΝΟΤΡΟΦΩΝ ΜΙΚΡΟΟΡΓΑΝΙΣΜΩΝ ΚΑΙ ΒΙΟΛΟΓΙΚΗ ΜΕΤΑΤΡΟΠΗ ΜΕΘΑΝΙΟΥ ΣΕ ΜΕΘΑΝΟΛΗ
 Σχολή Γεωτεχνικών Επιστημών και Διαχείρισης Περιβάλλοντος Πτυχιακή εργασία ΑΠΟΜΟΝΩΣΗ, ΤΑΥΤΟΠΟΙΗΣΗ ΜΕΘΑΝΟΤΡΟΦΩΝ ΜΙΚΡΟΟΡΓΑΝΙΣΜΩΝ ΚΑΙ ΒΙΟΛΟΓΙΚΗ ΜΕΤΑΤΡΟΠΗ ΜΕΘΑΝΙΟΥ ΣΕ ΜΕΘΑΝΟΛΗ Ιρένα Κυπριανίδου Λεμεσός, Μάιος
Σχολή Γεωτεχνικών Επιστημών και Διαχείρισης Περιβάλλοντος Πτυχιακή εργασία ΑΠΟΜΟΝΩΣΗ, ΤΑΥΤΟΠΟΙΗΣΗ ΜΕΘΑΝΟΤΡΟΦΩΝ ΜΙΚΡΟΟΡΓΑΝΙΣΜΩΝ ΚΑΙ ΒΙΟΛΟΓΙΚΗ ΜΕΤΑΤΡΟΠΗ ΜΕΘΑΝΙΟΥ ΣΕ ΜΕΘΑΝΟΛΗ Ιρένα Κυπριανίδου Λεμεσός, Μάιος
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
Supporting Information
 Supporting Information Montmorillonite KSF-Catalyzed One-pot, Three-component, Aza-Diels- Alder Reactions of Methylenecyclopropanes With Arylaldehydes and Aromatic Amines Li-Xiong Shao and Min Shi* General
Supporting Information Montmorillonite KSF-Catalyzed One-pot, Three-component, Aza-Diels- Alder Reactions of Methylenecyclopropanes With Arylaldehydes and Aromatic Amines Li-Xiong Shao and Min Shi* General
ΜΟΡΙΑ ΠΙΝΑΚΑ ΣΕΙΡΑ ΠΙΝΑΚΑ ΠΕΡΙΟΧΗ ΤΟΠΟΘΕΤΗΣΗΣ ΠΙΝΑΚΑΣ ΑΑ ΕΠΩΝΥΜΟ ΟΝΟΜΑ ΠΑΤΡΩΝΥΜΟ ΚΛΑΔΟΣ ΤΡΙΤΕΚΝΟ Σ ΔΙΕΥΘΥΝΣΗ ΕΚΠ/ΣΗΣ
 1 ΜΑΡΑΜΗ ΕΥΑΓΓΕΛΟ ΝΙΚΟΛΑΟ ΠΕ16.01 ΟΧΙ Β 1 38,715 Α Θεσσαλονίκης ΔΙΕΥΘΥΝΗ Π.Ε. ΘΕΑΛΟΝΙΚΗ Α 2 ΚΟΛΛΙΑ ΩΤΗΡΙΑ ΠΑΝΑΓΙΩΤΗ ΠΕ16.01 ΟΧΙ Β 2 17,29 Β Αθηνών ΔΙΕΥΘΥΝΗ Π.Ε. ΑΘΗΝΑ Β 3 ΔΕΠΟΤΗ ΩΤΗΡΙΟ ΚΩΝΤΑΝΤΙΝΟ ΠΕ16.01
1 ΜΑΡΑΜΗ ΕΥΑΓΓΕΛΟ ΝΙΚΟΛΑΟ ΠΕ16.01 ΟΧΙ Β 1 38,715 Α Θεσσαλονίκης ΔΙΕΥΘΥΝΗ Π.Ε. ΘΕΑΛΟΝΙΚΗ Α 2 ΚΟΛΛΙΑ ΩΤΗΡΙΑ ΠΑΝΑΓΙΩΤΗ ΠΕ16.01 ΟΧΙ Β 2 17,29 Β Αθηνών ΔΙΕΥΘΥΝΗ Π.Ε. ΑΘΗΝΑ Β 3 ΔΕΠΟΤΗ ΩΤΗΡΙΟ ΚΩΝΤΑΝΤΙΝΟ ΠΕ16.01
Strain gauge and rosettes
 Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Supporting Information. Asymmetric Binary-acid Catalysis with Chiral. Phosphoric Acid and MgF 2 : Catalytic
 Supporting Information Asymmetric Binary-acid Catalysis with Chiral Phosphoric Acid and MgF 2 : Catalytic Enantioselective Friedel-Crafts Reactions of β,γ- Unsaturated-α-Ketoesters Jian Lv, Xin Li, Long
Supporting Information Asymmetric Binary-acid Catalysis with Chiral Phosphoric Acid and MgF 2 : Catalytic Enantioselective Friedel-Crafts Reactions of β,γ- Unsaturated-α-Ketoesters Jian Lv, Xin Li, Long
Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια (MIS 365280)
 «Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
«Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
ΕΚΘΕΣΗ ΑΝΑΛΥΣΗΣ ΕΛΑΙΟΛΑ ΟΥ REPORT OF OLIVE OIL ANALYSIS
 ΕΘΝΙΚΗΣ ΑΝΤΙΣΤΑΣΕΩΣ 153, 2Ο2 ΟΟ ΚΙΑΤΟ ΚΟΡΙΝΘΙΑΣ, ΤΗΛ: 2742 Ο 22 554 FAX: 2742 Ο 22 545, E-MAIL: info@cadmion.gr, www.cadmion.gr 153, ETHNIKIS ANTISTASEOS STR., 2O2 OO KIATO KORINTHIA, GREECE, TEL: +3O
ΕΘΝΙΚΗΣ ΑΝΤΙΣΤΑΣΕΩΣ 153, 2Ο2 ΟΟ ΚΙΑΤΟ ΚΟΡΙΝΘΙΑΣ, ΤΗΛ: 2742 Ο 22 554 FAX: 2742 Ο 22 545, E-MAIL: info@cadmion.gr, www.cadmion.gr 153, ETHNIKIS ANTISTASEOS STR., 2O2 OO KIATO KORINTHIA, GREECE, TEL: +3O
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models
 Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
department listing department name αχχουντσ ϕανε βαλικτ δδσϕηασδδη σδηφγ ασκϕηλκ τεχηνιχαλ αλαν ϕουν διξ τεχηνιχαλ ϕοην µαριανι
 She selects the option. Jenny starts with the al listing. This has employees listed within She drills down through the employee. The inferred ER sttricture relates this to the redcords in the databasee
She selects the option. Jenny starts with the al listing. This has employees listed within She drills down through the employee. The inferred ER sttricture relates this to the redcords in the databasee
Γεωπονικό Πανεπιςτήμιο Αθηνών Τμήμα Αξιοποίηςησ Φυςικών Πόρων και Γεωργικήσ Μηχανικήσ
 Γεωπονικό Πανεπιςτήμιο Αθηνών Τμήμα Αξιοποίηςησ Φυςικών Πόρων και Γεωργικήσ Μηχανικήσ Εργαςτήριο Γεωργικών Καταςκευών ΠΜΣ Ενεργειακά Συςτήματα και Ανανεώςιμεσ Πηγέσ Ενέργειασ Διδακτορική διατριβή Καιιηέξγεηα
Γεωπονικό Πανεπιςτήμιο Αθηνών Τμήμα Αξιοποίηςησ Φυςικών Πόρων και Γεωργικήσ Μηχανικήσ Εργαςτήριο Γεωργικών Καταςκευών ΠΜΣ Ενεργειακά Συςτήματα και Ανανεώςιμεσ Πηγέσ Ενέργειασ Διδακτορική διατριβή Καιιηέξγεηα
DuPont Suva 95 Refrigerant
 Technical Information T-95 SI DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 SI DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
ΠΑΤΡΩΝΥΜΟ / ΟΝΟΜΑ ΣΥΖΥΓΟΥ 1 ΑΓΟΡΑΣΤΟΥ ΜΑΡΙΑ ΤΟΥ ΔΗΜΗΤΡΙΟΥ 2 ΑΘΑΝΑΣΙΑΔΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΠΑΥΛΟΥ 3 ΑΚΤΣΟΓΛΟΥ ΣΩΚΡΑΤΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ
 Υποψήφιοι ημοτικοί Σύμβουλοι: ΠΑΤΡΩΝΥΜΟ / ΣΥΖΥΓΟΥ 1 ΑΓΟΡΑΣΤΟΥ ΜΑΡΙΑ ΤΟΥ ΔΗΜΗΤΡΙΟΥ 2 ΑΘΑΝΑΣΙΑΔΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΠΑΥΛΟΥ 3 ΑΚΤΣΟΓΛΟΥ ΣΩΚΡΑΤΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 4 ΑΛΦΑΤΖΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 5 ΑΜΟΡΓΙΑΝΟΣ ΝΙΚΟΛΑΟΣ
Υποψήφιοι ημοτικοί Σύμβουλοι: ΠΑΤΡΩΝΥΜΟ / ΣΥΖΥΓΟΥ 1 ΑΓΟΡΑΣΤΟΥ ΜΑΡΙΑ ΤΟΥ ΔΗΜΗΤΡΙΟΥ 2 ΑΘΑΝΑΣΙΑΔΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΠΑΥΛΟΥ 3 ΑΚΤΣΟΓΛΟΥ ΣΩΚΡΑΤΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 4 ΑΛΦΑΤΖΗΣ ΙΩΑΝΝΗΣ ΤΟΥ ΓΕΩΡΓΙΟΥ 5 ΑΜΟΡΓΙΑΝΟΣ ΝΙΚΟΛΑΟΣ
Προσομοίωση BP με το Bizagi Modeler
 Προσομοίωση BP με το Bizagi Modeler Α. Τσαλγατίδου - Γ.-Δ. Κάπος Πρόγραμμα Μεταπτυχιακών Σπουδών Τεχνολογία Διοίκησης Επιχειρησιακών Διαδικασιών 2017-2018 BPMN Simulation with Bizagi Modeler: 4 Levels
Προσομοίωση BP με το Bizagi Modeler Α. Τσαλγατίδου - Γ.-Δ. Κάπος Πρόγραμμα Μεταπτυχιακών Σπουδών Τεχνολογία Διοίκησης Επιχειρησιακών Διαδικασιών 2017-2018 BPMN Simulation with Bizagi Modeler: 4 Levels
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
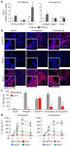 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
DuPont Suva 95 Refrigerant
 Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Friend or foe: differential responses of rice to invasion by mutualistic or. pathogenic fungi revealed by RNAseq and metabolite profiling
 Supplementary information Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling Xi-Hui Xu 1, Chen Wang 1, Shu-Xian Li
Supplementary information Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling Xi-Hui Xu 1, Chen Wang 1, Shu-Xian Li
Supplementary information:
 Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Διερεύνηση ακουστικών ιδιοτήτων Νεκρομαντείου Αχέροντα
 Διερεύνηση ακουστικών ιδιοτήτων Νεκρομαντείου Αχέροντα Βασίλειος Α. Ζαφρανάς Παναγιώτης Σ. Καραμπατζάκης ΠΕΡΙΛΗΨΗ H εργασία αφορά μία σειρά μετρήσεων του χρόνου αντήχησης της υπόγειας κρύπτης του «Νεκρομαντείου»
Διερεύνηση ακουστικών ιδιοτήτων Νεκρομαντείου Αχέροντα Βασίλειος Α. Ζαφρανάς Παναγιώτης Σ. Καραμπατζάκης ΠΕΡΙΛΗΨΗ H εργασία αφορά μία σειρά μετρήσεων του χρόνου αντήχησης της υπόγειας κρύπτης του «Νεκρομαντείου»
The Free Internet Journal for Organic Chemistry
 The Free Internet Journal for Organic Chemistry Paper Archive for Organic Chemistry Arkivoc 2018, part iii, S1-S6 Synthesis of dihydropyranones and dihydropyrano[2,3- d][1,3]dioxine-diones by cyclization
The Free Internet Journal for Organic Chemistry Paper Archive for Organic Chemistry Arkivoc 2018, part iii, S1-S6 Synthesis of dihydropyranones and dihydropyrano[2,3- d][1,3]dioxine-diones by cyclization
Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ. Σπίγγος Γεώργιος
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΤΜΗΜΑ ΑΓΡΟΝΟΜΩΝ ΤΟΠΟΓΡΑΦΩΝ ΜΗΧΑΝΙΚΩΝ ΤΟΜΕΑΣ ΤΟΠΟΓΡΑΦΙΑΣ-ΕΡΓΑΣΤΗΡΙΟ ΤΗΛΕΠΙΣΚΟΠΗΣΗΣ Αξιολόγηση των Φασματικού Διαχωρισμού στην Διάκριση Διαφορετικών Τύπων Εδάφους ΔΙΠΛΩΜΑΤΙΚΗ
The toxicity of three chitin synthesis inhibitors to Calliptamus italicus Othoptera Acridoidea
 2011 48 4 909 914 * 1 2** 2 2 2 2 2*** 1. 110161 2. 100081 026000 3 Calliptamus italicus L. 3 LC 50 LC 90 1. 34 14. 17 mg / L LC 50 LC 90 2. 09 45. 22 mg / L 50 mg / L 14 d 87% 100% 50 mg / L 50% The toxicity
2011 48 4 909 914 * 1 2** 2 2 2 2 2*** 1. 110161 2. 100081 026000 3 Calliptamus italicus L. 3 LC 50 LC 90 1. 34 14. 17 mg / L LC 50 LC 90 2. 09 45. 22 mg / L 50 mg / L 14 d 87% 100% 50 mg / L 50% The toxicity
 CSAE WPS/2009-06 Figure 1: Cut Flower Exports from Kenya, 1995-2007 Table 1: Firms in Areas with and w/out Conflict Panel A - Export Records Variable Observations Mean in No-Conflict
CSAE WPS/2009-06 Figure 1: Cut Flower Exports from Kenya, 1995-2007 Table 1: Firms in Areas with and w/out Conflict Panel A - Export Records Variable Observations Mean in No-Conflict
Homework 3 Solutions
 Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
Homework 3 Solutions Igor Yanovsky (Math 151A TA) Problem 1: Compute the absolute error and relative error in approximations of p by p. (Use calculator!) a) p π, p 22/7; b) p π, p 3.141. Solution: For
DuPont Suva. DuPont. Thermodynamic Properties of. Refrigerant (R-410A) Technical Information. refrigerants T-410A ENG
 Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
Supporting Information
 Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ & ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΜΕΛΕΤΗ ΚΑΙ ΚΑΤΑΣΚΕΥΗ ΙΑΤΑΞΗΣ ΜΕΤΡΗΣΗΣ ΘΕΡΜΟΧΩΡΗΤΙΚΟΤΗΤΑΣ ΣΕ ΜΑΓΝΗΤΙΚΟ ΠΕ ΙΟ ΓΙΑ ΧΑΡΑΚΤΗΡΙΣΜΟ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ & ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΜΕΛΕΤΗ ΚΑΙ ΚΑΤΑΣΚΕΥΗ ΙΑΤΑΞΗΣ ΜΕΤΡΗΣΗΣ ΘΕΡΜΟΧΩΡΗΤΙΚΟΤΗΤΑΣ ΣΕ ΜΑΓΝΗΤΙΚΟ ΠΕ ΙΟ ΓΙΑ ΧΑΡΑΚΤΗΡΙΣΜΟ
[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,
![[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13, [11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,](/thumbs/89/98694254.jpg) in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
ΜΔΛΔΣΖ ΔΝΓΟΣΡΑΥΤΝΖ Δ ΥΑΛΤΒΔ ΘΔΡΜΖ ΔΛΑΖ
 ΔΘΝΗΚΟ ΜΔΣΟΒΗΟ ΠΟΛΤΣΔΥΝΔΗΟ ΔΙΑΣΜΗΜΑΣΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΣΑΠΣΤΥΙΑΚΩΝ ΠΟΤΔΩΝ (Δ.Π.Μ..): "ΔΠΗΣΖΜΖ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΛΗΚΧΝ" ΜΔΛΔΣΖ ΔΝΓΟΣΡΑΥΤΝΖ Δ ΥΑΛΤΒΔ ΘΔΡΜΖ ΔΛΑΖ ΜΕΣΑΠΣΤΥΙΑΚΗ ΕΡΓΑΙΑ ΕΔΡΒΑ ΑΗΜΗΛΗΑΝΟ Διπλωματούτος
ΔΘΝΗΚΟ ΜΔΣΟΒΗΟ ΠΟΛΤΣΔΥΝΔΗΟ ΔΙΑΣΜΗΜΑΣΙΚΟ ΠΡΟΓΡΑΜΜΑ ΜΕΣΑΠΣΤΥΙΑΚΩΝ ΠΟΤΔΩΝ (Δ.Π.Μ..): "ΔΠΗΣΖΜΖ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΛΗΚΧΝ" ΜΔΛΔΣΖ ΔΝΓΟΣΡΑΥΤΝΖ Δ ΥΑΛΤΒΔ ΘΔΡΜΖ ΔΛΑΖ ΜΕΣΑΠΣΤΥΙΑΚΗ ΕΡΓΑΙΑ ΕΔΡΒΑ ΑΗΜΗΛΗΑΝΟ Διπλωματούτος
Effect of Genotype and Environment on Salvia miltiorrhiza Roots Using LC/MS Based Metabolomics
 S1 of S18 Effect of Genotype and Environment on Salvia miltiorrhiza Roots Using LC/MS Based Metabolomics Qi Zhao, Zhenqiao Song, Xinsheng Fang, Yuling Pan, Linlin Guo, Tian Liu and Jianhua Wang Table S1.
S1 of S18 Effect of Genotype and Environment on Salvia miltiorrhiza Roots Using LC/MS Based Metabolomics Qi Zhao, Zhenqiao Song, Xinsheng Fang, Yuling Pan, Linlin Guo, Tian Liu and Jianhua Wang Table S1.
SAMSUNG ELECTRONICS CO., LTD TEST REPORT SAMSUNG ELECTRONICS CO., LTD. 1, Samsung-Ro, Giheung-Gu, Yongin-Si, Gyeonggi-Do 17113, Korea
 SAMSUNG ELECTRONICS CO., LTD TEST REPORT Prepared For: SAMSUNG ELECTRONICS CO., LTD 1, Samsung-Ro, Giheung-Gu, Yongin-Si, Gyeonggi-Do 17113, Korea Product Name: LED Model Number: SPMWHX1228FXXXXXXXX Prepared
SAMSUNG ELECTRONICS CO., LTD TEST REPORT Prepared For: SAMSUNG ELECTRONICS CO., LTD 1, Samsung-Ro, Giheung-Gu, Yongin-Si, Gyeonggi-Do 17113, Korea Product Name: LED Model Number: SPMWHX1228FXXXXXXXX Prepared
1 h, , CaCl 2. pelamis) 58.1%, (Headspace solid -phase microextraction and gas chromatography -mass spectrometry,hs -SPME - Vol. 15 No.
 2 0 1 5 7 Journal of Chinese Institute of Food Science and Technology Vol. 15 No. 7 Jul. 2 0 1 5 1,2 1 1 1 1* ( 1 315211 2 315502) : ( ) : (E-Nose), - : GC-MS, 20.98% 2.5%, 53.15% 11.2%, 0. 51% 71.86%
2 0 1 5 7 Journal of Chinese Institute of Food Science and Technology Vol. 15 No. 7 Jul. 2 0 1 5 1,2 1 1 1 1* ( 1 315211 2 315502) : ( ) : (E-Nose), - : GC-MS, 20.98% 2.5%, 53.15% 11.2%, 0. 51% 71.86%
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Development of the Nursing Program for Rehabilitation of Woman Diagnosed with Breast Cancer
 Development of the Nursing Program for Rehabilitation of Woman Diagnosed with Breast Cancer Naomi Morota Newman M Key Words woman diagnosed with breast cancer, rehabilitation nursing care program, the
Development of the Nursing Program for Rehabilitation of Woman Diagnosed with Breast Cancer Naomi Morota Newman M Key Words woman diagnosed with breast cancer, rehabilitation nursing care program, the
ΤΜΗΜΑ ΕΚΠΑΙ ΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΣΥΝΕΧΙΖΟΜΕΝΗ ΕΚΠΑΙ ΕΥΣΗ ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΤΜΗΜΑ ΕΚΠΑΙ ΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΣΥΝΕΧΙΖΟΜΕΝΗ ΕΚΠΑΙ ΕΥΣΗ ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ ΙΕΡΕΥΝΗΣΗ ΤΩΝ ΕΠΙΜΟΡΦΩΤΙΚΩΝ ΑΝΑΓΚΩΝ ΤΩΝ ΣΥΜΒΟΥΛΩΝ ΨΥΧΟΛΟΓΩΝ ΤΩΝ ΣΧΟΛΕΙΩΝ
ΤΜΗΜΑ ΕΚΠΑΙ ΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΣΥΝΕΧΙΖΟΜΕΝΗ ΕΚΠΑΙ ΕΥΣΗ ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ ΙΕΡΕΥΝΗΣΗ ΤΩΝ ΕΠΙΜΟΡΦΩΤΙΚΩΝ ΑΝΑΓΚΩΝ ΤΩΝ ΣΥΜΒΟΥΛΩΝ ΨΥΧΟΛΟΓΩΝ ΤΩΝ ΣΧΟΛΕΙΩΝ
Current Status of PF SAXS beamlines. 07/23/2014 Nobutaka Shimizu
 Current Status of PF SAXS beamlines 07/23/2014 Nobutaka Shimizu BL-6A Detector SAXS:PILATUS3 1M WAXD:PILATUS 100K Wavelength 1.5Å (Fix) Camera Length 0.25, 0.5, 1.0, 2.0, 2.5 m +WAXD Chamber:0.75, 1.0,
Current Status of PF SAXS beamlines 07/23/2014 Nobutaka Shimizu BL-6A Detector SAXS:PILATUS3 1M WAXD:PILATUS 100K Wavelength 1.5Å (Fix) Camera Length 0.25, 0.5, 1.0, 2.0, 2.5 m +WAXD Chamber:0.75, 1.0,
ΜΕΛΕΤΗ ΑΝΤΙΔΡΑΣΗΣ ΕΣΤΕΡΟΠΟΊΗΣΗΣ ΓΑΛΑΚΤΙΚΟΥ ΟΞΕΟΣ ΜΕ ΑΙΘΑΝΟΛΗ ΓΙΑ ΠΑΡΑΓΩΓΗ ΓΑΛΑΚΤΙΚΟΥ ΑΙΘΥΛΕΣΤΕΡΑ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΜΕΛΕΤΗ ΑΝΤΙΔΡΑΣΗΣ ΕΣΤΕΡΟΠΟΊΗΣΗΣ ΓΑΛΑΚΤΙΚΟΥ ΟΞΕΟΣ ΜΕ ΑΙΘΑΝΟΛΗ ΓΙΑ ΠΑΡΑΓΩΓΗ ΓΑΛΑΚΤΙΚΟΥ ΑΙΘΥΛΕΣΤΕΡΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΜΕΛΕΤΗ ΑΝΤΙΔΡΑΣΗΣ ΕΣΤΕΡΟΠΟΊΗΣΗΣ ΓΑΛΑΚΤΙΚΟΥ ΟΞΕΟΣ ΜΕ ΑΙΘΑΝΟΛΗ ΓΙΑ ΠΑΡΑΓΩΓΗ ΓΑΛΑΚΤΙΚΟΥ ΑΙΘΥΛΕΣΤΕΡΑ
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ
 ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
Protease-catalysed Direct Asymmetric Mannich Reaction in Organic Solvent
 Supplementary information for the paper Protease-catalysed Direct Asymmetric Mannich Reaction in Organic Solvent Yang Xue, Ling-Po Li, Yan-Hong He * & Zhi Guan * School of Chemistry and Chemical Engineering,
Supplementary information for the paper Protease-catalysed Direct Asymmetric Mannich Reaction in Organic Solvent Yang Xue, Ling-Po Li, Yan-Hong He * & Zhi Guan * School of Chemistry and Chemical Engineering,
Ανάκτηση Πληροφορίας
 Ανάκτηση Πληροφορίας Αποτίμηση Αποτελεσματικότητας Μέτρα Απόδοσης Precision = # σχετικών κειμένων που επιστρέφονται # κειμένων που επιστρέφονται Recall = # σχετικών κειμένων που επιστρέφονται # συνολικών
Ανάκτηση Πληροφορίας Αποτίμηση Αποτελεσματικότητας Μέτρα Απόδοσης Precision = # σχετικών κειμένων που επιστρέφονται # κειμένων που επιστρέφονται Recall = # σχετικών κειμένων που επιστρέφονται # συνολικών
(Biomass utilization for electric energy production)
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ T.Ε.I. ΠΕΙΡΑΙΑ ΣΧΟΛΗ: ΤΕΧΝΟΛΟΓΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΤΜΗΜΑ: ΗΛΕΚΤΡΟΛΟΓΙΑΣ Επιβλέπων: ΠΕΤΡΟΣ Γ. ΒΕΡΝΑΔΟΣ, Ομότιμος Καθηγητής Συνεπιβλέπουσα: ΕΡΙΕΤΤΑ Ι. ΖΟΥΝΤΟΥΡΙΔΟΥ, Παν. Υπότροφος
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ T.Ε.I. ΠΕΙΡΑΙΑ ΣΧΟΛΗ: ΤΕΧΝΟΛΟΓΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΤΜΗΜΑ: ΗΛΕΚΤΡΟΛΟΓΙΑΣ Επιβλέπων: ΠΕΤΡΟΣ Γ. ΒΕΡΝΑΔΟΣ, Ομότιμος Καθηγητής Συνεπιβλέπουσα: ΕΡΙΕΤΤΑ Ι. ΖΟΥΝΤΟΥΡΙΔΟΥ, Παν. Υπότροφος
HOMEWORK 4 = G. In order to plot the stress versus the stretch we define a normalized stretch:
 HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
Jesse Maassen and Mark Lundstrom Purdue University November 25, 2013
 Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
Notes on Average Scattering imes and Hall Factors Jesse Maassen and Mar Lundstrom Purdue University November 5, 13 I. Introduction 1 II. Solution of the BE 1 III. Exercises: Woring out average scattering
