Supplementary Online Content
|
|
|
- Καλλίας Μιχαλολιάκος
- 7 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, doi: /jamaoncol etable 1. Clinical characteristics of FAP and patients etable 2. Pathology characteristics of FAP and polyps etable 3. Immune profile of polyps compared to FAP etable 4. Immune profile of FAP polyps polyps, and carcinomas etable 5. Gene set enrichment analysis of FAP and polyps, tumor etable 6. Mutational rate analysis in FAP, and hypermutated polyps, and and hypermutated Stage I-II TCGA CRC using RNAseq data etable 7. Immune profile of polyps classified by mutational rate (hyper- versus nonhypermutant) etable 8. Predicted Class I and II HLA genotype of FAP and normal mucosa samples etable 9. List of Predicted neoantigens etable 10. Pathways analysis of MHC I and II neoantigens etable 11. Germline and somatic mutation detected in FAP and polyps. Mutational data derived from RNA-seq analysis efigure 1. Immune profile of Lynch syndrome premalignancy and carcinomas efigure 2. Correlation of DNA and RNA mutation rate in samples from TCGA efigure 3. Mutation Spectrum of FAP and polyps efigure 4. Immunohistochemical staining of CD4 and FOXP3 efigure 5. MHC Class I and II neoantigens in Lynch syndrome and FAP premalignancy, and carcinomas ematerials and Methods ereferences
2 This supplementary material has been provided by the authors to give readers additional information about their work.
3 etable 1. Clinical characteristics of FAP and patients Patient ID Gender Age Race Col ore ctal Sx No. of polyp s detect ed Cancer Dx Germline mutation Gene gdna Protein No. of normal mucosa sequen ced No. of adenom as sequen ced FAP_1 M 35 W IRA >100 APC c.1880dupa p.ala630* FAP_2 M 33 W IRA <5 APC c.3810t>a p.cys1270* FAP_3 F 42 W IRA 5 APC del exon 14 p.? FAP_4 M 42 W P >100 HCC, APC c.622c>t p.gln208* D FAP_5 F 40 W IRA 5 APC del exons 8-9 p.? FAP_6 M 37 W IRA <5 APC c.1658g>a p.trp553* FAP_7 M 65 W IRA <50 APC c.477c>g p.tyr159* FAP_8 F 25 AA NP >100 D APC c.4733_4734del p.cys1578tyrf s*12 FAP_9 F 28 W IRA >100 EC, APC c.847c>t p.arg283* OC FAP_10 F 25 W P >100 APC c.3810t>a p.cys1270* _1 M 53 W RH 1 CC MSH6 c.3744_3773del3 p.his1248_ser del _2 F 46 W NP 3 EC MSH2 c.687dela p.ala230leufs* _3 M 52 W LH 1 CC PMS2 del exon 14 p.? _4 F 37 W NP 1 MSH2 c.1034g>a p.trp345* _5 F 53 W NP 1 MSH2 c g>a p.? _6 M 58 W NP 2 MSH2 del exons 1-6 p.? _7 M 76 W LH 2 CC MSH2 c.1216c>t p.arg406* _8 F 68 W NP 4 EC MSH6 c.3238_3239delc p.leu1080valfs T *12 _9 F 62 W NP 1 EC MSH6 c.3860ins4 p.p1289x _10 F 43 W RH 3 CC MSH6 c.2645_2653delt TAAGTCTA p.phe882* No. of tumors sequen ced
4 _11 F 63 W TC 3 CC, EC MSH6 c.3699_3702dela p.lys1233asnf R GAA s*6 _12 F 35 W SC 0 CC MLH1 c.1279c>t p.gln427* _13 F 76 W SC 0 CC, MLH BC, GIST c.1918c>t p.pro640ser _14 F 61 W RH 0 CC PMS2 c.2059c>t p.arg687trp Abbreviations: M, male; F, Female; W, white; AA, African-American; IRA, ileorectal anastomosis; P, pouch; RH, right hemicolectomy; LH, left hemicolectomy; TCR, transverse colon resection; SC, subtotal colectomy; NP, not performed; NA, not available; CC, colon cancer; HCC, hepatocellular carcinoma; HepBI, hepatoblastoma; D, desmoid; EC, endometrial cancer; OC, ovarian cancer. *Denotes Hispanic ancestry.
5 etable 2. Pathology characteristics of FAP and polyps Patient ID Sample ID Disease Pathology Location Size/TNM IHC-MSH2 IHC-MSH6 IHC-MLH1 IHC-PMS2 FAP_1 FAP_G2 FAP TA R 5-10 NA NA NA NA FAP_1 FAP_G3 FAP TA R 5-10 NA NA NA NA FAP_2 FAP_G53 FAP TA R <5 NA NA NA NA FAP_2 FAP_G55 FAP TA R <5 NA NA NA NA FAP_3 FAP_G5 FAP TA R <5 NA NA NA NA FAP_3 FAP_G6 FAP TA R <5 NA NA NA NA FAP_4 FAP_G13 FAP TA R 5-10 NA NA NA NA FAP_4 FAP_G14 FAP TA R 5-10 NA NA NA NA FAP_4 FAP_G15 FAP TA R 5-10 NA NA NA NA FAP_5 FAP_G25 FAP TA R <5 NA NA NA NA FAP_6 FAP_G29 FAP TA R NA NA NA NA NA FAP_7 FAP_G39 FAP TA R <5 NA NA NA NA FAP_8 FAP_G47 FAP TA R <5 NA NA NA NA FAP_8 FAP_G48 FAP TA R <5 NA NA NA NA FAP_9 FAP_G67 FAP TA R 5-10 NA NA NA NA FAP_10 FAP_G61 FAP TA NA NA NA NA NA NA _1 _EBL1 TA D 5-10 Intact Intact NA NA _2 _EBL3 TA A <5 Intact Intact NA NA _3 _EBL5 HP D 5-10 NA NA Intact Intact _4 _EBL7 TA with HGD D >10 Lost Lost NA NA _5 _EBL9 TA R 5-10 Lost Lost NA NA _6 _EBL11 TA A >10 Lost Lost NA NA _7 _EBL13 TA A 5-10 Intact Intact NA NA _8 _EBL18 HP D 5-10 Intact Intact NA NA _9 _EBL26 TA D 5-10 NA Intact NA NA _10 _EBL20 TA D 5-10 Intact Lost NA NA _11 _EBL23 TA A <5 Intact Intact NA NA _12 _T3 CA D pt3n0 Intact Intact Lost Lost _13 _T2 CA D pt4an0 Intact Intact Lost Lost _14 _T1 CA A pt2n0 Intact Intact Intact Lost Abbreviations: TA, tubular adenoma; HP, hyperplastic polyp; HGD, high grade dysplasia; A, cecum/ascending/hepatic flexure; D, transverse/descending/sigmoid; R, rectum/rectosigmoid; NA, not assessed. Assessment of expression of MMR proteins in polyps was based on the already known germline mutations.therefore, only the protein pair matching the germline mutation was performed.
6 etable 3. Immune profile of polyps compared to FAP Gene Mean FAP Mean t test FDR t test P value CD IFNG TBX CD8A GZMB PRF IL CTLA PDCD LAG CD IL17A RORC IL23A FOXP IL TGFB PTGS IL1B IL IL IL12A TNF IDO NOS HIF1A Genes linked to the immune microenvironment of CRC and MMR deficiency were grouped by lineage and/or function (Th1/Tc1, CTL, Th17, Treg, proinflammation, and metabolism) as previously reported by Llosa et al, Cancer Discovery (2016). Expression values are counts per million. The statistical tests were performed on log2-transformed CPM. Welch's t-test and multiple correction by Benjamini & Hochberg were used (FDR<0.05). Significantly different genes are in bold
7 etable 4. Immune profile of FAP polyps polyps, and carcinomas Gene Mean Tumor Mean Mean FAP Tumor vs FDR vs FAP FDR Tumor vs FAP FDR CD IFNG TBX CD8A GZMB PRF IL IL17A RORC IL23A FOXP IL TGFB PTGS IL1B IL IL IL12A TNF CTLA PDCD LAG CD IDO NOS HIF1A Genes linked to the immune microenvironment of CRC and MMR deficiency were grouped by lineage and/or function (Th1/Tc1, CTL, Th17, Treg, proinflammation, and metabolism) as previously reported by Llosa et al, Cancer Discovery (2016). Expression values are expressed in counts per million. The statistical tests were performed on log2-transformed CPM. Significance between each groups is tested using ANOVA and Tukey s Test, and Benjamini & Hochberg is used for multiple correction (FDR<0.05). Significantly different genes are in bold.
8 etable 5. Gene set enrichment analysis of FAP and polyps, tumor Pathways FAP vs FDR FAP vs Tumor FDR vs Tumor FDR Number of genes Reference IMMUNE_ESTIMATE PMID_ IMMUNE_RESP_GO_BP GO_BP IMMUNE_TH1_GALON PMID_ _ IMMUNE_TH17_GOUNARI PMID_ IMMUNE_TREG_LUCAS PMID_ PD1_REACTOME REACTOME T-cell-signature PMID_ Significance between each groups is tested using ANOVA and Tukey s Test, and Benjamini & Hochberg is used for multiple correction (FDR<0.05). Significantly different genes are in bold. etable 6. Mutational rate analysis in FAP, and hypermutated polyps, and and hypermutated Stage I-II TCGA CRC using RNAseq data Sample ID Type of Sample Mutations/Mb _EBL _EBL _EBL _EBL _EBL _EBL _EBL _EBL _EBL11 Hyper 33.2 _EBL20 Hyper _EBL9 Hyper _T1 Tumor 21.4 _T2 Tumor 26.4 _T3 Tumor 51.5 FAP_G13 FAP
9 FAP_G14 FAP FAP_G15 FAP FAP_G2 FAP FAP_G25 FAP FAP_G29 FAP FAP_G3 FAP FAP_G39 FAP FAP_G47 FAP FAP_G48 FAP FAP_G5 FAP FAP_G53 FAP FAP_G55 FAP FAP_G6 FAP FAP_G61 FAP FAP_G67 FAP TCGA-AA-3517 CRC Stage I,II TCGA-AA-3524 CRC Stage I,II TCGA-AA-3526 CRC Stage I,II TCGA-AA-3527 CRC Stage I,II TCGA-AA-3530 CRC Stage I,II TCGA-AA-3531 CRC Stage I,II TCGA-AA-3532 CRC Stage I,II TCGA-AA-3534 CRC Stage I,II TCGA-AA-3538 CRC Stage I,II TCGA-AA-3544 CRC Stage I,II TCGA-AA-3549 CRC Stage I,II TCGA-AA-3556 CRC Stage I,II TCGA-AA-3561 CRC Stage I,II TCGA-AA-3664 CRC Stage I,II TCGA-AA-3667 CRC Stage I,II TCGA-AA-3673 CRC Stage I,II TCGA-AA-3685 CRC Stage I,II TCGA-AA-3812 CRC Stage I,II TCGA-AA-3814 CRC Stage I,II TCGA-AA-3818 CRC Stage I,II 6.398
10 TCGA-AA-3831 CRC Stage I,II TCGA-AA-3837 CRC Stage I,II TCGA-AA-3850 CRC Stage I,II TCGA-AA-3852 CRC Stage I,II 5.49 TCGA-AA-3854 CRC Stage I,II TCGA-AA-3856 CRC Stage I,II TCGA-AA-3858 CRC Stage I,II TCGA-AA-3866 CRC Stage I,II TCGA-AA-3875 CRC Stage I,II TCGA-AA-3939 CRC Stage I,II TCGA-AA-3956 CRC Stage I,II TCGA-AA-3975 CRC Stage I,II TCGA-AA-3980 CRC Stage I,II TCGA-AA-A004 CRC Stage I,II TCGA-AA-A00Z CRC Stage I,II 5.55 TCGA-AA-A017 CRC Stage I,II TCGA-AA-A024 CRC Stage I,II TCGA-AA-A029 CRC Stage I,II TCGA-AA-A02W CRC Stage I,II TCGA-AA-A02Y CRC Stage I,II TCGA-AY-4071 CRC Stage I,II TCGA-AA-3710 CRC Stage I,II Hyper TCGA-AA-3715 CRC Stage I,II Hyper TCGA-AA-3864 CRC Stage I,II Hyper TCGA-AA-3877 CRC Stage I,II Hyper TCGA-AA-A00N CRC Stage I,II Hyper TCGA-AA-A010 CRC Stage I,II Hyper 59.93
11 etable 7. Immune profile of polyps classified by mutational rate (hyper- versus non-hypermutant) Gene Hypermutant Mean muta nt Mean Hypervs muta nt FDR Hypervs muta nt p-value CD IFNG TBX CD8A GZMB PRF IL IL17A RORC IL23A FOXP IL TGFB PTGS IL1B IL IL IL12A TNF CTLA PDCD LAG CD IDO NOS
12 etable 8. Predicted Class I and II HLA genotype of FAP and normal mucosa samples Patie nt ID FAP_ 1 FAP_ 4 FAP_ 5 FAP_ 6 FAP_ 3 FAP_ 7 FAP_ 8 FAP_ 2 FAP_ 9 FAP_ 10 _5 Sample ID FAP_G 1 FAP_G 16 FAP_G 26 FAP_G 31 FAP_G 4 FAP_G 40 FAP_G 49 FAP_G 54 FAP_G 60 FAP_G 62 _EBL 10 Region A Region B Region C Region DQA Region DQB Region DRB Allele Allele Allele Allele Allele Allele Allele 1 Allele 2 Allele 1 Allele 2 Allele 1 Allele HLA- HLA- HLA- HLA- HLA- HLA- DQA1*01 DQA1*01 DQB1*06 DQB1*05 DRB1*01 DRB1*15 A*01: A*11: B*39: B*52: C*12: C*12: :02 :02 :01 :01 :01 : HLA- A*25: 01 HLA- A*29: 02 HLA- A*01: 01 HLA- A*24: 02 HLA- A*29: 02 HLA- A*34: 02 HLA- A*33: 03 HLA- A*24: 02 HLA- A*24: 02 HLA- A*23: 01 HLA- A*11: 01 HLA- A*24: 02 HLA- A*24: 02 HLA- A*02: 01 HLA- A*03: 01 HLA- A*02: 01 HLA- A*03: 01 HLA- A*02: 01 HLA- A*02: 01 HLA- A*01: 01 HLA- B*18: 01 HLA- B*56: 01 HLA- B*15: 17 HLA- B*18: 01 HLA- B*44: 03 HLA- B*45: 01 HLA- B*44: 03 HLA- B*18: 01 HLA- B*15: 07 HLA- B*44: 03 HLA- B*35: 01 HLA- B*51: 01 HLA- B*07: 02 HLA- B*07: 02 HLA- B*07: 02 HLA- B*53: 01 HLA- B*51: 01 HLA- B*44: 02 HLA- B*15: 07 HLA- B*07: 02 HLA- C*04: 01 HLA- C*02: 02 HLA- C*07: 01 HLA- C*05: 01 HLA- C*16: 01 HLA- C*04: 01 HLA- C*15: 02 HLA- C*05: 01 HLA- C*03: 04 HLA- C*04: 01 HLA- C*12: 03 HLA- C*01: 02 HLA- C*07: 01 HLA- C*07: 02 HLA- C*07: 02 HLA- C*16: 01 HLA- C*07: 01 HLA- C*12: 03 HLA- C*03: 04 HLA- C*07: 02 DQA1*01 :04 DQA1*02 :01 DQA1*01 :02 DQA1*03 :02 DQA1*02 :01 DQA1*04 :01 DQA1*01 :02 DQA1*03 :02 DQA1*03 :02 DQA1*05 :01 DQA1*01 :04 DQA1*01 :02 DQA1*01 :02 DQA1*02 :01 DQA1*01 :02 DQA1*01 :02 DQA1*02 :01 DQA1*05 :01 DQA1*01 :02 DQA1*01 :01 DQB1*06 :13 DQB1*05 :01 DQB1*06 :13 DQB1*03 :02 DQB1*06 :02 DQB1*06 :02 DQB1*05 :02 DQB1*03 :01 DQB1*06 :11 DQB1*03 :01 DQB1*05 :01 DQB1*02 :01 DQB1*06 :13 DQB1*02 :01 DQB1*02 :01 DQB1*03 :09 DQB1*02 :05 DQB1*03 :01 DQB1*03 :02 DQB1*05 :01 DRB1*01 :01 DRB1*01 :01 DRB1*13 :02 DRB1*04 :01 DRB1*07 :01 DRB1*15 :03 DRB1*16 :01 DRB1*04 :01 DRB1*04 :04 DRB1*01 :01 DRB1*15 :01 DRB1*07 :01 DRB1*15 :01 DRB1*07 :01 DRB1*15 :01 DRB1*08 :04 DRB1*07 :01 DRB1*11 :04 DRB1*15 :01 DRB1*11 :04
13 etable 9. List of Predicted Neoantigens Chromosome Position Sample Gene Best.MT.Score Expression Allele MHC (TPM) Frequency FAP_G13 PRDM MHC_I FAP_G13 SPEN MHC_I FAP_G13 POGK MHC_I FAP_G13 CEP MHC_I FAP_G13 TRMT1L MHC_I FAP_G13 SH3PXD2A MHC_I FAP_G13 TCF7L MHC_I FAP_G13 DCLRE1A MHC_I FAP_G13 ARHGAP MHC_I FAP_G13 RECQL MHC_I FAP_G13 RPAP MHC_I FAP_G13 LRIG MHC_I FAP_G13 USP MHC_I FAP_G13 MIPEP MHC_I FAP_G13 NYNRIN MHC_I FAP_G13 HECTD MHC_I FAP_G13 HEATR5A MHC_I FAP_G13 EFCAB MHC_I FAP_G13 UBE3A MHC_I FAP_G13 HERC MHC_I FAP_G13 FANCI MHC_I FAP_G13 COTL MHC_I FAP_G13 NCOR MHC_I FAP_G13 NCOR MHC_I FAP_G13 TSEN MHC_I FAP_G13 PIAS MHC_I FAP_G13 RDH MHC_I
14 FAP_G13 ZNF MHC_I FAP_G13 EIF2AK MHC_I FAP_G13 BMPR MHC_I FAP_G13 BMPR MHC_I FAP_G13 BMPR MHC_I FAP_G13 BMPR MHC_I FAP_G13 ARFGAP MHC_I FAP_G13 MINA MHC_I FAP_G13 OPA MHC_I FAP_G13 ATP13A MHC_I FAP_G13 SEC24B MHC_I FAP_G13 DDX MHC_I FAP_G13 PAPD MHC_I FAP_G13 IL6ST MHC_I FAP_G13 TMEM161B MHC_I FAP_G13 AIM MHC_I FAP_G13 CDC MHC_I FAP_G13 PHF MHC_I FAP_G13 VPS MHC_I FAP_G13 FIGNL MHC_I FAP_G13 STEAP MHC_I FAP_G13 MEPCE MHC_I FAP_G13 NBN MHC_I FAP_G13 NFIL MHC_I FAP_G14 ZMYM MHC_I FAP_G14 AHCTF MHC_I FAP_G14 CSTF2T MHC_I FAP_G14 ZNF518A MHC_I FAP_G14 BBIP MHC_I FAP_G14 TCF7L MHC_I FAP_G14 CACUL MHC_I
15 FAP_G14 PLEKHA MHC_I FAP_G14 PEX MHC_I FAP_G14 SPI MHC_I FAP_G14 CDC42BPG MHC_I FAP_G14 DHCR MHC_I FAP_G14 DLAT MHC_I FAP_G14 LRP MHC_I FAP_G14 LRP MHC_I FAP_G14 FBXO MHC_I FAP_G14 MPHOSPH MHC_I FAP_G14 MAP4K MHC_I FAP_G14 PTGR MHC_I FAP_G14 UBE3A MHC_I FAP_G14 TJP MHC_I FAP_G14 SLC12A MHC_I FAP_G14 EFTUD MHC_I FAP_G14 HEATR MHC_I FAP_G14 CWC MHC_I FAP_G14 ANKRD MHC_I FAP_G14 DUSP MHC_I FAP_G14 CTSA MHC_I FAP_G14 CSE1L MHC_I FAP_G14 TRANK MHC_I FAP_G14 WDR MHC_I FAP_G14 MINA MHC_I FAP_G14 MYLK MHC_I FAP_G14 OPA MHC_I FAP_G14 IL6ST MHC_I FAP_G14 TMEM161B MHC_I FAP_G14 STEAP MHC_I FAP_G14 MTERF MHC_I
16 FAP_G14 NBN MHC_I FAP_G15 H6PD MHC_I FAP_G15 USP MHC_I FAP_G15 PDE3B MHC_I FAP_G15 SUV420H MHC_I FAP_G15 PGM2L MHC_I FAP_G15 SORL MHC_I FAP_G15 FBXL MHC_I FAP_G15 PRIM MHC_I FAP_G15 HSPH MHC_I FAP_G15 SLC7A MHC_I FAP_G15 TC2N MHC_I FAP_G15 PARN MHC_I FAP_G15 NOMO MHC_I FAP_G15 EARS MHC_I FAP_G15 RSPRY MHC_I FAP_G15 ASNA MHC_I FAP_G15 ZNF MHC_I FAP_G15 ROCK MHC_I FAP_G15 PID MHC_I FAP_G15 TRIP MHC_I FAP_G15 MKKS MHC_I FAP_G15 ATXN MHC_I FAP_G15 MINA MHC_I FAP_G15 NMNAT MHC_I FAP_G15 PLOD MHC_I FAP_G15 EHHADH MHC_I FAP_G15 SEC24B MHC_I FAP_G15 FHDC MHC_I FAP_G15 LMBRD MHC_I FAP_G15 ITGA MHC_I
17 FAP_G15 DHX MHC_I FAP_G15 IL6ST MHC_I FAP_G15 ANKRA MHC_I FAP_G15 MED MHC_I FAP_G15 AIM MHC_I FAP_G15 CDC MHC_I FAP_G15 REPS MHC_I FAP_G15 KATNA MHC_I FAP_G15 CASD MHC_I FAP_G15 TRIM MHC_I FAP_G15 WASH MHC_I FAP_G15 CAAP MHC_I FAP_G15 EXD MHC_I X FAP_G15 ABCB MHC_I X FAP_G15 NDUFA MHC_I X FAP_G15 AIFM MHC_I FAP_G2 MRPL MHC_I FAP_G2 DEPDC MHC_I FAP_G2 ZNF MHC_I FAP_G2 NUP MHC_I FAP_G2 AHCTF MHC_I FAP_G2 KCNMA MHC_I FAP_G2 CCSER MHC_I FAP_G2 XPOT MHC_I FAP_G2 UBE3B MHC_I FAP_G2 CDK MHC_I FAP_G2 HAUS MHC_I FAP_G2 RALGAPA MHC_I FAP_G2 UBN MHC_I FAP_G2 POLR3E MHC_I FAP_G2 METTL MHC_I
18 FAP_G2 SOX MHC_I FAP_G2 REV MHC_I FAP_G2 RIF MHC_I FAP_G2 STAM MHC_I FAP_G2 GPCPD MHC_I FAP_G2 RPRD1B MHC_I FAP_G2 CLASP MHC_I FAP_G2 CLASP MHC_I FAP_G2 SEC22C MHC_I FAP_G2 GBE MHC_I FAP_G2 GPR MHC_I FAP_G2 TBCK MHC_I FAP_G2 LARP1B MHC_I FAP_G2 CSNK1A MHC_I FAP_G2 ATG MHC_I FAP_G2 RPF MHC_I FAP_G2 TRIM MHC_I FAP_G2 LMBR MHC_I X FAP_G2 IDS MHC_I FAP_G25 ZNF MHC_I FAP_G25 SLC11A MHC_I FAP_G25 WBP MHC_I FAP_G25 RPS17L MHC_I FAP_G25 ATP6V0D MHC_I FAP_G25 BAIAP MHC_I FAP_G25 ACTR MHC_I FAP_G25 CD MHC_I FAP_G25 GALNT MHC_I FAP_G25 SLC41A MHC_I FAP_G25 ETFDH MHC_I FAP_G25 ATG MHC_I
19 FAP_G29 PLEKHA MHC_I FAP_G29 WAPAL MHC_I FAP_G29 FRA10AC MHC_I FAP_G29 TAF1D MHC_I FAP_G29 PARP MHC_I FAP_G29 CHURC MHC_I FAP_G29 TC2N MHC_I FAP_G29 PDIA MHC_I FAP_G29 NCL MHC_I FAP_G29 CSTF MHC_I FAP_G29 IFT MHC_I FAP_G29 ATXN MHC_I FAP_G29 SLC39A MHC_I FAP_G29 HNRNPH MHC_I FAP_G29 ADAMDEC MHC_I FAP_G29 SET MHC_I X FAP_G29 BEX MHC_I FAP_G3 ZMYM MHC_I FAP_G3 ELOVL MHC_I FAP_G3 FAF MHC_I FAP_G3 UBE2Q MHC_I FAP_G3 SUV420H MHC_I FAP_G3 PPP6R MHC_I FAP_G3 RALGAPA MHC_I FAP_G3 TRIP MHC_I FAP_G3 BTBD MHC_I FAP_G3 C15orf MHC_I FAP_G3 HERC MHC_I FAP_G3 TRIP MHC_I FAP_G3 LAMA MHC_I FAP_G3 ZNF MHC_I
20 FAP_G3 RDH MHC_I FAP_G3 ANO MHC_I FAP_G3 SLCO2A MHC_I FAP_G3 ANKHD MHC_I FAP_G3 ANKHD MHC_I EIF4EBP FAP_G3 TBP MHC_I FAP_G3 SAMD9L MHC_I FAP_G3 PPAPDC1B MHC_I FAP_G3 RMI MHC_I X FAP_G3 BTK MHC_I FAP_G39 API MHC_I FAP_G39 FIBP MHC_I FAP_G39 CEP MHC_I FAP_G39 ERGIC MHC_I FAP_G39 SNX MHC_I FAP_G39 PDXDC MHC_I FAP_G39 THRA MHC_I FAP_G39 UBXN MHC_I FAP_G39 FARSB MHC_I FAP_G39 IGLL MHC_I FAP_G39 RPP MHC_I FAP_G39 OPA MHC_I FAP_G39 DHX MHC_I FAP_G39 FAM105A MHC_I FAP_G39 CWC MHC_I FAP_G39 UBTD MHC_I FAP_G39 ACAT MHC_I X FAP_G39 NONO MHC_I FAP_G47 EPB MHC_I FAP_G47 EFCAB MHC_I FAP_G47 MCOLN MHC_I
21 FAP_G47 FMO MHC_I FAP_G47 IRF MHC_I FAP_G47 PCGF MHC_I FAP_G47 ZFP MHC_I FAP_G47 PCF MHC_I FAP_G47 GOLT1B MHC_I FAP_G47 PUS7L MHC_I FAP_G47 TBC1D MHC_I FAP_G47 DLGAP MHC_I FAP_G47 SIPA1L MHC_I FAP_G47 CCNB MHC_I FAP_G47 CNOT MHC_I FAP_G47 ATP6V0D MHC_I FAP_G47 TEFM MHC_I FAP_G47 THOC MHC_I FAP_G47 PPP4R MHC_I FAP_G47 SLC39A MHC_I FAP_G47 GTF2F MHC_I FAP_G47 PRKD MHC_I FAP_G47 PCBP MHC_I FAP_G47 MRPS MHC_I FAP_G47 PKP MHC_I FAP_G47 C3orf MHC_I FAP_G47 SKIL MHC_I FAP_G47 NUP MHC_I FAP_G47 PLK MHC_I FAP_G47 FBXO MHC_I FAP_G47 SKIV2L MHC_I FAP_G47 FEM1C MHC_I FAP_G47 M MHC_I FAP_G47 LMBR MHC_I
22 FAP_G48 RERE MHC_I FAP_G48 FAF MHC_I FAP_G48 USP MHC_I FAP_G48 EPC MHC_I FAP_G48 PHLDA MHC_I FAP_G48 ZFP MHC_I FAP_G48 PCF MHC_I FAP_G48 NXPE MHC_I FAP_G48 APPL MHC_I FAP_G48 CLIP MHC_I FAP_G48 DTD MHC_I FAP_G48 ZNF MHC_I FAP_G48 FOPNL MHC_I FAP_G48 VPS MHC_I FAP_G48 CNOT MHC_I FAP_G48 ATP6V0D MHC_I FAP_G48 ESRP MHC_I FAP_G48 ABCA MHC_I FAP_G48 THOC MHC_I FAP_G48 PIGN MHC_I FAP_G48 ZBTB7A MHC_I FAP_G48 PEPD MHC_I FAP_G48 PTPRH MHC_I FAP_G48 PUM MHC_I FAP_G48 EIF5B MHC_I FAP_G48 CCDC MHC_I FAP_G48 WNK MHC_I FAP_G5 EIF4G MHC_I FAP_G5 KIAA0319L MHC_I FAP_G5 NRD MHC_I FAP_G5 UBAP2L MHC_I
23 FAP_G5 ELF MHC_I FAP_G5 FAM21B MHC_I FAP_G5 USP MHC_I FAP_G5 ADD MHC_I FAP_G5 TTC MHC_I FAP_G5 PRPF MHC_I FAP_G5 SMARCD MHC_I FAP_G5 USP MHC_I FAP_G5 TBK MHC_I FAP_G5 GNS MHC_I FAP_G5 ANKRD13A MHC_I FAP_G5 FBXO MHC_I FAP_G5 MTMR MHC_I FAP_G5 RMDN MHC_I FAP_G5 KIAA MHC_I FAP_G5 DHX MHC_I FAP_G5 ARHGAP MHC_I FAP_G5 MYO18A MHC_I FAP_G5 EZH MHC_I FAP_G5 PIAS MHC_I FAP_G5 ZADH MHC_I FAP_G5 PTBP MHC_I FAP_G5 DNMT MHC_I FAP_G5 ROCK MHC_I FAP_G5 PPM1B MHC_I FAP_G5 USP MHC_I FAP_G5 CDCA MHC_I FAP_G5 PLCB MHC_I FAP_G5 PAXBP MHC_I FAP_G5 OXSR MHC_I FAP_G5 FAM208A MHC_I
24 FAP_G5 SMC MHC_I FAP_G5 SMC MHC_I FAP_G5 MFN MHC_I FAP_G5 PPP3CA MHC_I FAP_G5 TBCK MHC_I FAP_G5 KIAA MHC_I FAP_G5 FNIP MHC_I FAP_G5 ELOVL MHC_I FAP_G5 RAD MHC_I FAP_G5 C5orf MHC_I FAP_G5 LRRC16A MHC_I FAP_G5 PAQR MHC_I FAP_G5 SESN MHC_I FAP_G5 TRIM MHC_I FAP_G5 ERMP MHC_I X FAP_G5 THOC MHC_I FAP_G53 RERE MHC_I FAP_G53 ODF2L MHC_I FAP_G53 PLEKHO MHC_I FAP_G53 SUCO MHC_I FAP_G53 SUCO MHC_I FAP_G53 MIA MHC_I FAP_G53 FAM171A MHC_I FAP_G53 ARHGAP MHC_I FAP_G53 ZNF33A MHC_I FAP_G53 GPRIN MHC_I FAP_G53 NUP MHC_I FAP_G53 ERGIC MHC_I FAP_G53 IPO MHC_I FAP_G53 R3HDM MHC_I FAP_G53 MIS18BP MHC_I
25 FAP_G53 UBE3A MHC_I FAP_G53 DUOXA MHC_I FAP_G53 MYO1E MHC_I FAP_G53 CLPX MHC_I FAP_G53 ZC3H7A MHC_I FAP_G53 GLG MHC_I FAP_G53 MIS MHC_I FAP_G53 MIS MHC_I FAP_G53 MED MHC_I FAP_G53 EMC MHC_I FAP_G53 RBM MHC_I FAP_G53 STK MHC_I FAP_G53 SBF MHC_I FAP_G53 DALRD MHC_I FAP_G53 ABHD MHC_I FAP_G53 SLC41A MHC_I FAP_G53 ABTB MHC_I FAP_G53 FAM175A MHC_I FAP_G53 KIF13A MHC_I FAP_G53 PEX MHC_I FAP_G53 SLC17A MHC_I FAP_G53 BCKDHB MHC_I FAP_G53 ZHX MHC_I FAP_G55 WDTC MHC_I FAP_G55 ODF2L MHC_I FAP_G55 PLEKHO MHC_I FAP_G55 TTC MHC_I FAP_G55 ARHGAP MHC_I FAP_G55 NUP MHC_I FAP_G55 ERGIC MHC_I FAP_G55 XPOT MHC_I
26 FAP_G55 SART MHC_I FAP_G55 RALGAPA MHC_I FAP_G55 STAT MHC_I FAP_G55 FNIP MHC_I FAP_G55 C5orf MHC_I FAP_G55 FAM135A MHC_I FAP_G55 SLC17A MHC_I FAP_G55 SAMD MHC_I FAP_G55 TRIM MHC_I FAP_G55 IARS MHC_I FAP_G6 EXOSC MHC_I FAP_G6 NRD MHC_I FAP_G6 ODF2L MHC_I FAP_G6 WARS MHC_I FAP_G6 NBPF MHC_I FAP_G6 ISG20L MHC_I FAP_G6 DDX MHC_I FAP_G6 USP MHC_I FAP_G6 ZBED MHC_I FAP_G6 TTC MHC_I FAP_G6 CDC42BPG MHC_I FAP_G6 PPFIA MHC_I FAP_G6 STK38L MHC_I FAP_G6 USP MHC_I FAP_G6 ATP2B MHC_I FAP_G6 ANKRD13A MHC_I FAP_G6 ZC3H7A MHC_I FAP_G6 RRN MHC_I FAP_G6 POLR3E MHC_I FAP_G6 DHX MHC_I FAP_G6 TERF2IP MHC_I
27 FAP_G6 MYO18A MHC_I FAP_G6 SLC38A MHC_I FAP_G6 ZNF MHC_I FAP_G6 FCGBP MHC_I FAP_G6 ROCK MHC_I FAP_G6 EIF2AK MHC_I FAP_G6 PPM1B MHC_I FAP_G6 EIF2AK MHC_I FAP_G6 PCED1A MHC_I FAP_G6 FERMT MHC_I FAP_G6 TTLL MHC_I FAP_G6 TOP2B MHC_I FAP_G6 OXSR MHC_I FAP_G6 STAG MHC_I FAP_G6 MFN MHC_I FAP_G6 UGT2A MHC_I FAP_G6 FRG MHC_I FAP_G6 SLC30A MHC_I FAP_G6 SLC12A MHC_I FAP_G6 CFTR MHC_I FAP_G6 TMEM MHC_I FAP_G6 ERMP MHC_I FAP_G6 KDM4C MHC_I FAP_G6 SMC MHC_I FAP_G6 TMEM MHC_I X FAP_G6 THOC MHC_I FAP_G61 OMA MHC_I FAP_G61 SMYD MHC_I FAP_G61 YME1L MHC_I FAP_G61 AHNAK MHC_I FAP_G61 PPFIA MHC_I
28 FAP_G61 EED MHC_I FAP_G61 CCDC MHC_I FAP_G61 LRIG MHC_I FAP_G61 MED13L MHC_I FAP_G61 COG MHC_I FAP_G61 PYGL MHC_I FAP_G61 DYNC1H MHC_I FAP_G61 BNIP MHC_I FAP_G61 CDK MHC_I FAP_G61 SPOP MHC_I FAP_G61 TRIM MHC_I FAP_G61 AXIN MHC_I FAP_G61 MOB3A MHC_I FAP_G61 SH3GL MHC_I FAP_G61 DPP MHC_I FAP_G61 FCGBP MHC_I FAP_G61 THAP MHC_I FAP_G61 PLAGL MHC_I FAP_G61 NCOA MHC_I FAP_G61 YTHDF MHC_I FAP_G61 TCAIM MHC_I FAP_G61 CEP MHC_I FAP_G61 ATR MHC_I FAP_G61 PLOD MHC_I FAP_G61 SNX MHC_I FAP_G61 KDM3B MHC_I FAP_G61 RGL MHC_I FAP_G61 ASCC MHC_I FAP_G61 MICAL MHC_I FAP_G61 TRIM MHC_I FAP_G61 NUP MHC_I
29 FAP_G61 TMUB MHC_I FAP_G67 SCMH MHC_I FAP_G67 FMO MHC_I FAP_G67 MANSC MHC_I FAP_G67 PYGL MHC_I FAP_G67 GNPNAT MHC_I FAP_G67 TRIM MHC_I FAP_G67 ORC MHC_I FAP_G67 SF3B MHC_I FAP_G67 SACM1L MHC_I FAP_G67 PCBP MHC_I FAP_G67 UBA MHC_I FAP_G67 CDC MHC_I FAP_G67 AHCYL MHC_I FAP_G67 TLE MHC_I FAP_G67 C9orf MHC_I X FAP_G67 UBE2A MHC_I _EBL1 ZBTB MHC_I _EBL1 PDSS MHC_I _EBL1 ACY MHC_I _EBL1 LPCAT MHC_I _EBL1 AQR MHC_I _EBL1 AQR MHC_I _EBL1 VWA MHC_I _EBL1 GEMIN MHC_I _EBL1 VAT MHC_I _EBL1 EFTUD MHC_I _EBL1 NPEPPS MHC_I _EBL1 NPEPPS MHC_I _EBL1 HOXB MHC_I _EBL1 ATP13A MHC_I
30 _EBL1 ZNF MHC_I _EBL1 PTPRH MHC_I _EBL1 PARP MHC_I _EBL1 FBXO MHC_I _EBL1 FAF MHC_I _EBL1 SESN MHC_I _EBL1 INTS MHC_I _EBL1 TLE MHC_I _EBL1 SECISBP MHC_I _EBL1 XPA MHC_I X _EBL1 ARSE MHC_I X _EBL1 PSMD MHC_I _EBL11 GPR MHC_I _EBL11 EPHA MHC_I _EBL11 HSPG MHC_I _EBL11 EPB MHC_I _EBL11 ZMYM MHC_I _EBL11 PLK MHC_I _EBL11 CYP2J MHC_I _EBL11 SRSF MHC_I _EBL11 POGZ MHC_I _EBL11 DENND4B MHC_I _EBL11 DEDD MHC_I _EBL11 B4GALT MHC_I _EBL11 C4BPB MHC_I _EBL11 DMBT MHC_I _EBL11 ATHL MHC_I _EBL11 NUP MHC_I _EBL11 CTSF MHC_I _EBL11 PGM2L MHC_I _EBL11 PRSS MHC_I
31 _EBL11 NOP MHC_I _EBL11 TBX MHC_I _EBL11 COL4A MHC_I _EBL11 SLC7A MHC_I _EBL11 NFATC MHC_I _EBL11 PTPN MHC_I _EBL11 PACS MHC_I _EBL11 SCAMP MHC_I _EBL11 NLRC MHC_I _EBL11 PLEKHG MHC_I _EBL11 GLG MHC_I _EBL11 DEF MHC_I _EBL11 GIT MHC_I _EBL11 HNF1B MHC_I _EBL11 KAT2A MHC_I _EBL11 STAT5A MHC_I _EBL11 HGS MHC_I _EBL11 C17orf MHC_I _EBL11 C19orf MHC_I _EBL11 MYO9B MHC_I _EBL11 PIK3R MHC_I _EBL11 DMPK MHC_I _EBL11 NAPA MHC_I _EBL11 ZNF MHC_I _EBL11 ZNF MHC_I _EBL11 RMND5A MHC_I _EBL11 THNSL MHC_I _EBL11 SLC9A MHC_I _EBL11 AGAP MHC_I _EBL11 PCMTD MHC_I _EBL11 MAPK MHC_I
32 _EBL11 BCR MHC_I _EBL11 SMARCB MHC_I _EBL11 MPST MHC_I _EBL11 LDOC1L MHC_I _EBL11 CAMK MHC_I _EBL11 GPR MHC_I _EBL11 ABTB MHC_I _EBL11 EIF4A MHC_I _EBL11 NELFA MHC_I _EBL11 SRD5A MHC_I _EBL11 UGT MHC_I _EBL11 TMEM MHC_I _EBL11 PCDH MHC_I _EBL11 LARP MHC_I _EBL11 ECI MHC_I _EBL11 HIST1H2BK MHC_I _EBL11 VARS MHC_I _EBL11 TUBE MHC_I _EBL11 NCOA MHC_I _EBL11 TRRAP MHC_I _EBL11 AGAP MHC_I _EBL11 HR MHC_I _EBL11 SLC25A MHC_I _EBL11 RNF MHC_I _EBL11 MTSS MHC_I _EBL11 TOP1MT MHC_I _EBL11 EEF1D MHC_I _EBL11 CPSF MHC_I _EBL11 KIAA MHC_I _EBL11 UBAP MHC_I _EBL11 TMEM8B MHC_I
33 _EBL11 GNE MHC_I _EBL11 ZBTB MHC_I _EBL11 NCBP MHC_I _EBL11 PTPN MHC_I _EBL11 NTMT MHC_I _EBL11 TSC MHC_I X _EBL11 MED MHC_I X _EBL11 CASK MHC_I X _EBL11 HDAC MHC_I X _EBL11 AIFM MHC_I _EBL13 UCHL MHC_I _EBL13 PEX MHC_I _EBL13 NDN MHC_I _EBL13 DDA MHC_I _EBL13 ANKRD MHC_I _EBL13 VRK MHC_I _EBL13 APOL MHC_I _EBL13 SBF MHC_I _EBL13 CPT1B MHC_I _EBL13 NICN MHC_I _EBL13 TPRA MHC_I _EBL13 C6orf MHC_I _EBL13 C9orf MHC_I _EBL18 SMPD MHC_I _EBL18 CPT1A MHC_I _EBL18 TM9SF MHC_I _EBL18 ATP6V0D MHC_I _EBL18 RBKS MHC_I _EBL18 MEMO MHC_I _EBL18 KLF MHC_I _EBL20 RPRD MHC_I
34 _EBL20 ZNF MHC_I _EBL20 ASPM MHC_I _EBL20 CENPF MHC_I _EBL20 NRBF MHC_I _EBL20 SIRT MHC_I _EBL20 SCD MHC_I _EBL20 MOB MHC_I _EBL20 KBTBD MHC_I _EBL20 MAP3K MHC_I _EBL20 ACY MHC_I _EBL20 POLD MHC_I _EBL20 CTN MHC_I _EBL20 ADCY MHC_I _EBL20 SLC11A MHC_I _EBL20 LRP MHC_I _EBL20 NTN MHC_I _EBL20 OAS MHC_I _EBL20 MTMR MHC_I _EBL20 SLITRK MHC_I _EBL20 ARHGAP MHC_I _EBL20 DNAAF MHC_I _EBL20 DCAF MHC_I _EBL20 ELMSAN MHC_I _EBL20 SPATA MHC_I _EBL20 AQR MHC_I _EBL20 MYO5C MHC_I _EBL20 TCF MHC_I _EBL20 TBC1D2B MHC_I _EBL20 GFER MHC_I _EBL20 MT MHC_I _EBL20 RRN MHC_I
35 _EBL20 TMC MHC_I _EBL20 ZG MHC_I _EBL20 GPR MHC_I _EBL20 ZCCHC MHC_I _EBL20 CENPV MHC_I _EBL20 DHX MHC_I _EBL20 TLK MHC_I _EBL20 PSMC MHC_I _EBL20 HID MHC_I _EBL20 LLGL MHC_I _EBL20 BAIAP MHC_I _EBL20 ADNP MHC_I _EBL20 PVRL MHC_I _EBL20 ZNF MHC_I _EBL20 DDX MHC_I _EBL20 ADCY MHC_I _EBL20 PNPT MHC_I _EBL20 USP MHC_I _EBL20 NDUFS MHC_I _EBL20 SGPP MHC_I _EBL20 MFF MHC_I _EBL20 FASTKD MHC_I _EBL20 C20orf MHC_I _EBL20 FAM83D MHC_I _EBL20 ATP9A MHC_I _EBL20 UCKL MHC_I _EBL20 PI4KA MHC_I _EBL20 SETD MHC_I _EBL20 DALRD MHC_I _EBL20 HYAL MHC_I _EBL20 ACTR MHC_I
36 _EBL20 PROS MHC_I _EBL20 ZBTB MHC_I _EBL20 NELFA MHC_I _EBL20 NIPAL MHC_I _EBL20 FRYL MHC_I _EBL20 KIAA MHC_I _EBL20 PPID MHC_I _EBL20 ING MHC_I _EBL20 TRAPPC MHC_I _EBL20 SRP MHC_I _EBL20 PGGT1B MHC_I _EBL20 TMEM63B MHC_I _EBL20 SOD MHC_I _EBL20 CCZ1B MHC_I _EBL20 MEST MHC_I _EBL20 XPO MHC_I _EBL20 INTS MHC_I _EBL20 DDHD MHC_I _EBL20 NBN MHC_I _EBL20 SPAG MHC_I _EBL20 CPSF MHC_I _EBL20 SECISBP MHC_I _EBL20 TGFBR MHC_I _EBL20 RNF MHC_I _EBL20 NSMF MHC_I X _EBL20 TRMT2B MHC_I X _EBL20 TRMT2B MHC_I _EBL23 DDAH MHC_I _EBL23 AMPD MHC_I _EBL23 FGFR MHC_I _EBL23 BATF MHC_I
37 _EBL23 TBRG MHC_I _EBL23 RNFT MHC_I _EBL23 WDR MHC_I _EBL23 MST1R MHC_I _EBL23 GALNT MHC_I _EBL26 KIFAP MHC_I _EBL26 TUBGCP MHC_I _EBL26 SBF MHC_I _EBL26 KLRB MHC_I _EBL26 EGLN MHC_I _EBL26 AMFR MHC_I _EBL26 ATP6V0D MHC_I _EBL26 ATP13A MHC_I _EBL26 ATP13A MHC_I _EBL26 ATG9A MHC_I _EBL26 CRNKL MHC_I _EBL26 PBRM MHC_I _EBL26 FAM208A MHC_I _EBL26 PLD MHC_I _EBL26 ATP8A MHC_I _EBL26 TTC MHC_I _EBL26 MCM MHC_I _EBL26 KIAA MHC_I X _EBL26 ARSE MHC_I X _EBL26 DIAPH MHC_I X _EBL26 TMLHE MHC_I _EBL3 DUSP MHC_I _EBL3 PTPRR MHC_I _EBL3 MTERFD MHC_I _EBL3 CPSF MHC_I _EBL3 XPO MHC_I
38 _EBL3 ATXN2L MHC_I _EBL3 ATP6V0D MHC_I _EBL3 ADORA2B MHC_I _EBL3 ERAL MHC_I _EBL3 TEX MHC_I _EBL3 LIPG MHC_I _EBL3 SGTA MHC_I _EBL3 CCDC MHC_I _EBL3 ARHGEF MHC_I _EBL3 ZNF MHC_I _EBL3 PTCD MHC_I _EBL3 LMAN2L MHC_I _EBL3 TOPBP MHC_I _EBL3 SH3D MHC_I _EBL3 RWDD MHC_I _EBL3 SKP MHC_I _EBL3 VARS MHC_I _EBL3 RRP MHC_I _EBL3 MED MHC_I _EBL3 TRIM MHC_I X _EBL3 MTM MHC_I _EBL5 USP MHC_I _EBL5 SMARCD MHC_I _EBL5 TBL MHC_I _EBL5 CWC MHC_I _EBL5 PLOD MHC_I _EBL5 TBC1D MHC_I _EBL7 POLL MHC_I _EBL7 ZFC3H MHC_I _EBL7 CRY MHC_I _EBL7 OAS MHC_I
39 _EBL7 ATP6V0A MHC_I _EBL7 BTBD MHC_I _EBL7 AQR MHC_I _EBL7 PEAK MHC_I _EBL7 KIAA MHC_I _EBL7 STAT5A MHC_I _EBL7 PLCD MHC_I _EBL7 COIL MHC_I _EBL7 MFSD MHC_I _EBL7 TBCD MHC_I _EBL7 ZBTB7A MHC_I _EBL7 MTA MHC_I _EBL7 RNF MHC_I _EBL7 KIAA MHC_I _EBL7 DLG MHC_I _EBL7 TLR MHC_I _EBL7 FAT MHC_I _EBL7 CCDC MHC_I _EBL7 MYO MHC_I _EBL7 FAM8A MHC_I _EBL7 C6orf MHC_I _EBL7 FBXL MHC_I _EBL7 INTS MHC_I _EBL7 FTSJ MHC_I _EBL7 PHF MHC_I _EBL7 WWP MHC_I _EBL7 FBXW MHC_I _EBL7 SETX MHC_I X _EBL7 ELF MHC_I _EBL9 ACAP MHC_I _EBL9 ATAD3A MHC_I
40 _EBL9 CDK11B MHC_I _EBL9 SKI MHC_I _EBL9 MTHFR MHC_I _EBL9 VPS13D MHC_I _EBL9 PLEKHM MHC_I _EBL9 EPHA MHC_I _EBL9 HP1BP MHC_I _EBL9 USP MHC_I _EBL9 WASF MHC_I _EBL9 YARS MHC_I _EBL9 RNF19B MHC_I _EBL9 ZMYM MHC_I _EBL9 IPO MHC_I _EBL9 MAST MHC_I _EBL9 HENMT MHC_I _EBL9 PTGFRN MHC_I _EBL9 VPS MHC_I _EBL9 THEM MHC_I _EBL9 XPR MHC_I _EBL9 PLXNA MHC_I _EBL9 EPHX MHC_I _EBL9 RBM MHC_I _EBL9 NID MHC_I _EBL9 NID MHC_I _EBL9 ARHGAP MHC_I _EBL9 DNAJC MHC_I _EBL9 ZNF518A MHC_I _EBL9 C10orf MHC_I _EBL9 SFXN MHC_I _EBL9 MCMBP MHC_I _EBL9 EPS8L MHC_I
41 _EBL9 ZNF MHC_I _EBL9 SLC43A MHC_I _EBL9 SYT MHC_I _EBL9 MARK MHC_I _EBL9 CDC42BPG MHC_I _EBL9 EHD MHC_I _EBL9 VPS MHC_I _EBL9 SIPA MHC_I _EBL9 PITPNM MHC_I _EBL9 ARAP MHC_I _EBL9 SIK MHC_I _EBL9 DPAGT MHC_I _EBL9 KDM5A MHC_I _EBL9 TNFRSF1A MHC_I _EBL9 ITPR MHC_I _EBL9 PKP MHC_I _EBL9 DGKA MHC_I _EBL9 TIMELESS MHC_I _EBL9 MON MHC_I _EBL9 VPS33A MHC_I _EBL9 RNF MHC_I _EBL9 RBM MHC_I _EBL9 TEP MHC_I _EBL9 ACIN MHC_I _EBL9 NYNRIN MHC_I _EBL9 PRKCH MHC_I _EBL9 ACTN MHC_I _EBL9 YLPM MHC_I _EBL9 ACYP MHC_I _EBL9 DICER MHC_I _EBL9 SPG MHC_I
42 _EBL9 SECISBP2L MHC_I _EBL9 TCF MHC_I _EBL9 BNIP MHC_I _EBL9 KIAA MHC_I _EBL9 TMEM8A MHC_I _EBL9 RAB11FIP MHC_I _EBL9 HAGH MHC_I _EBL9 TSC MHC_I _EBL9 PKD MHC_I _EBL9 TXNDC MHC_I _EBL9 MVP MHC_I _EBL9 E2F MHC_I _EBL9 ACSF MHC_I _EBL9 ATP2A MHC_I _EBL9 ZZEF MHC_I _EBL9 SPNS MHC_I _EBL9 MINK MHC_I _EBL9 TNK MHC_I _EBL9 MD MHC_I _EBL9 C17orf MHC_I _EBL9 EVI2B MHC_I _EBL9 ZNF MHC_I _EBL9 MYO MHC_I _EBL9 ERBB MHC_I _EBL9 DHX MHC_I _EBL9 WNK MHC_I _EBL9 PLEKHM MHC_I _EBL9 RNFT MHC_I _EBL9 APPBP MHC_I _EBL9 TMC MHC_I _EBL9 EPB41L MHC_I
43 _EBL9 POLRMT MHC_I _EBL9 WDR MHC_I _EBL9 MIDN MHC_I _EBL9 CSNK1G MHC_I _EBL9 STAP MHC_I _EBL9 C19orf MHC_I _EBL9 RAVER MHC_I _EBL9 CALR MHC_I _EBL9 TRMT MHC_I _EBL9 NACC MHC_I _EBL9 PKN MHC_I _EBL9 MAU MHC_I _EBL9 ANKRD MHC_I _EBL9 FOXA MHC_I _EBL9 NOSIP MHC_I _EBL9 ZNF MHC_I _EBL9 HSPBP MHC_I _EBL9 NOL MHC_I _EBL9 ZFP36L MHC_I _EBL9 PSME MHC_I _EBL9 MTIF MHC_I _EBL9 USP MHC_I _EBL9 GCFC MHC_I _EBL9 TMEM185B MHC_I _EBL9 PKP MHC_I _EBL9 PPIG MHC_I _EBL9 TRIP MHC_I _EBL9 KIF3B MHC_I _EBL9 NECAB MHC_I _EBL9 TRPC4AP MHC_I _EBL9 PKIG MHC_I
44 _EBL9 ADNP MHC_I _EBL9 GART MHC_I _EBL9 ABCG MHC_I _EBL9 YDJC MHC_I _EBL9 PPM1F MHC_I _EBL9 MYH MHC_I _EBL9 DDX MHC_I _EBL9 TUBGCP MHC_I _EBL9 PLXNB MHC_I _EBL9 FANCD MHC_I _EBL9 NUP MHC_I _EBL9 PVRL MHC_I _EBL9 PVRL MHC_I _EBL9 SPICE MHC_I _EBL9 MRPL MHC_I _EBL9 HLTF MHC_I _EBL9 FNDC3B MHC_I _EBL9 DVL MHC_I _EBL9 EPHB MHC_I _EBL9 DGKQ MHC_I _EBL9 MFSD MHC_I _EBL9 FRYL MHC_I _EBL9 BMP2K MHC_I _EBL9 ALPK MHC_I _EBL9 EXOSC MHC_I _EBL9 FAT MHC_I _EBL9 PDCD MHC_I _EBL9 6-Mar MHC_I _EBL9 NIPBL MHC_I _EBL9 ELOVL MHC_I _EBL9 ANKRA MHC_I
45 _EBL9 GFM MHC_I _EBL9 WDR MHC_I _EBL9 DHFR MHC_I _EBL9 ELL MHC_I _EBL9 CDC42SE MHC_I _EBL9 CCNG MHC_I _EBL9 RMND5B MHC_I _EBL9 NELFE MHC_I _EBL9 C6orf MHC_I _EBL9 KCNK MHC_I _EBL9 PEX MHC_I _EBL9 PEX MHC_I _EBL9 MRPS18A MHC_I _EBL9 FBXL MHC_I _EBL9 RMND MHC_I _EBL9 SNX MHC_I _EBL9 DPY19L MHC_I _EBL9 TBRG MHC_I _EBL9 BAZ1B MHC_I _EBL9 DTX MHC_I _EBL9 ACHE MHC_I _EBL9 CASP MHC_I _EBL9 ABCB MHC_I _EBL9 SLC39A MHC_I _EBL9 KAT6A MHC_I _EBL9 CHD MHC_I _EBL9 ASPH MHC_I _EBL9 STAU MHC_I _EBL9 ST3GAL MHC_I _EBL9 PTK MHC_I _EBL9 OPLAH MHC_I
46 _EBL9 CBWD MHC_I _EBL9 KIAA MHC_I _EBL9 KIAA MHC_I _EBL9 MLLT MHC_I _EBL9 TRPM MHC_I _EBL9 IARS MHC_I _EBL9 XPA MHC_I _EBL9 GALNT MHC_I _EBL9 KLF MHC_I _EBL9 STOM MHC_I _EBL9 SLC25A MHC_I _EBL9 DNM MHC_I _EBL9 NUP MHC_I _EBL9 NUP MHC_I _EBL9 RAPGEF MHC_I X _EBL9 PRKX MHC_I X _EBL9 STS MHC_I X _EBL9 BCOR MHC_I X _EBL9 P MHC_I X _EBL9 SLC6A MHC_I X _EBL9 BCAP MHC_I _T1 AGRN MHC_I _T1 MTOR MHC_I _T1 POMGNT MHC_I _T1 SMG MHC_I _T1 HSD17B MHC_I _T1 EPRS MHC_I _T1 GLUD MHC_I _T1 NUP MHC_I _T1 ILK MHC_I _T1 SBF MHC_I
47 _T1 TTC9C MHC_I _T1 ZFC3H MHC_I _T1 APAF MHC_I _T1 ANAPC MHC_I _T1 POLE MHC_I _T1 TPP MHC_I _T1 TPP MHC_I _T1 TPP MHC_I _T1 TFDP MHC_I _T1 ZFP36L MHC_I _T1 AHSA MHC_I _T1 CLCN MHC_I _T1 SRRM MHC_I _T1 USP MHC_I _T1 BOLA MHC_I _T1 STX MHC_I _T1 NLRC MHC_I _T1 RFWD MHC_I _T1 ARRB MHC_I _T1 NCOR MHC_I _T1 RNF MHC_I _T1 MOCOS MHC_I _T1 PLD MHC_I _T1 TOMM MHC_I _T1 BIRC MHC_I _T1 EPAS MHC_I _T1 EIF5B MHC_I _T1 ERCC MHC_I _T1 NOSTRIN MHC_I _T1 NOP MHC_I _T1 SEC23B MHC_I
48 _T1 PYGB MHC_I _T1 SON MHC_I _T1 CABIN MHC_I _T1 ACO MHC_I _T1 TCF MHC_I _T1 TADA MHC_I _T1 NBEAL MHC_I _T1 UBA MHC_I _T1 PVRL MHC_I _T1 PARP MHC_I _T1 TOPBP MHC_I _T1 ZNF MHC_I _T1 PAM MHC_I _T1 IRF MHC_I _T1 RARS MHC_I _T1 TBC1D MHC_I _T1 ITPR MHC_I _T1 BCKDHB MHC_I _T1 PEX MHC_I _T1 PLOD MHC_I _T1 EXT MHC_I _T1 TRAPPC MHC_I _T1 EPPK MHC_I X _T1 ATRX MHC_I _T2 EPHA MHC_I _T2 C1QA MHC_I _T2 HDAC MHC_I _T2 JUN MHC_I _T2 GBP MHC_I _T2 NBPF MHC_I _T2 DHX MHC_I
49 _T2 TPR MHC_I _T2 HNRNPU MHC_I _T2 PTEN MHC_I _T2 FAR MHC_I _T2 CAPRIN MHC_I _T2 ST MHC_I _T2 USP MHC_I _T2 DDX MHC_I _T2 USP MHC_I _T2 CHD MHC_I _T2 MAP4K MHC_I _T2 SIPA1L MHC_I _T2 DT MHC_I _T2 SMEK MHC_I _T2 VRK MHC_I _T2 HERC MHC_I _T2 TMEM87A MHC_I _T2 SMG MHC_I _T2 XPO MHC_I _T2 PHKB MHC_I _T2 ATP6V0D MHC_I _T2 CIRH1A MHC_I _T2 ANKRD MHC_I _T2 ACADVL MHC_I _T2 MPDU MHC_I _T2 CHD MHC_I _T2 PRPSAP MHC_I _T2 UTP MHC_I _T2 GSDMB MHC_I _T2 ATP6V0A MHC_I _T2 GRN MHC_I
50 _T2 KPNB MHC_I _T2 PSMD MHC_I _T2 BPTF MHC_I _T2 DSC MHC_I _T2 DYM MHC_I _T2 AP3D MHC_I _T2 NDUFA MHC_I _T2 ASNA MHC_I _T2 SPINT MHC_I _T2 SUPT5H MHC_I _T2 AKT MHC_I _T2 RAB MHC_I _T2 MSH MHC_I _T2 SNRNP MHC_I _T2 MMADHC MHC_I _T2 NDUFS MHC_I _T2 ACSL MHC_I _T2 PARVB MHC_I _T2 MLH MHC_I _T2 WDR MHC_I _T2 MUC MHC_I _T2 TMEM MHC_I _T2 G3BP MHC_I _T2 LRBA MHC_I _T2 FAT MHC_I _T2 TRIO MHC_I _T2 NIPBL MHC_I _T2 PIK3R MHC_I _T2 DCBLD MHC_I _T2 TRRAP MHC_I _T2 MUC MHC_I
51 _T2 MCM MHC_I _T2 SMARCA MHC_I _T2 DCTN MHC_I _T2 VPS13A MHC_I _T2 GLE MHC_I _T2 TUBB4B MHC_I X _T2 ACOT MHC_I X _T2 TAF MHC_I _T3 RERE MHC_I _T3 VPS13D MHC_I _T3 C1orf MHC_I _T3 BSDC MHC_I _T3 ZFYVE MHC_I _T3 PKN MHC_I _T3 DPYD MHC_I _T3 RFX MHC_I _T3 ADIPOR MHC_I _T3 LBR MHC_I _T3 ACBD MHC_I _T3 ALDH18A MHC_I _T3 GBF MHC_I _T3 BUB MHC_I _T3 ANO MHC_I _T3 MUC5B MHC_I _T3 MUC5B MHC_I _T3 PIK3C2A MHC_I _T3 CTNND MHC_I _T3 VPS MHC_I _T3 SYVN MHC_I _T3 KIAA MHC_I _T3 CASP MHC_I
52 _T3 SRPR MHC_I _T3 ATN MHC_I _T3 PFKM MHC_I _T3 R3HDM MHC_I _T3 CAND MHC_I _T3 PIBF MHC_I _T3 TPP MHC_I _T3 ACIN MHC_I _T3 AP1G MHC_I _T3 REC MHC_I _T3 YLPM MHC_I _T3 SPTLC MHC_I _T3 WARS MHC_I _T3 DYNC1H MHC_I _T3 EIF2AK MHC_I _T3 MGA MHC_I _T3 UACA MHC_I _T3 NDE MHC_I _T3 DCTPP MHC_I _T3 VPS MHC_I _T3 PIEZO MHC_I _T3 CHMP1A MHC_I _T3 PRPF MHC_I _T3 POLR2A MHC_I _T3 CHD MHC_I _T3 AP2B MHC_I _T3 MED MHC_I _T3 WNK MHC_I _T3 WNK MHC_I _T3 DHX MHC_I _T3 LRRC MHC_I
53 _T3 SPAG MHC_I _T3 TSEN MHC_I _T3 LLGL MHC_I _T3 FASN MHC_I _T3 SLC16A MHC_I _T3 PIK3C MHC_I _T3 AP3D MHC_I _T3 EEF MHC_I _T3 MCOLN MHC_I _T3 MYO1F MHC_I _T3 SMARCA MHC_I _T3 DOCK MHC_I _T3 ECSIT MHC_I _T3 TNPO MHC_I _T3 ZNF MHC_I _T3 SIPA1L MHC_I _T3 FCGBP MHC_I _T3 ARHGEF MHC_I _T3 DMPK MHC_I _T3 TMC MHC_I _T3 BIRC MHC_I _T3 FAM98A MHC_I _T3 LRPPRC MHC_I _T3 EPAS MHC_I _T3 PSME MHC_I _T3 GCC MHC_I _T3 RIF MHC_I _T3 GPD MHC_I _T3 PPIG MHC_I _T3 NCKAP MHC_I _T3 DIS3L MHC_I
54 _T3 PCED1A MHC_I _T3 PANK MHC_I _T3 KIF16B MHC_I _T3 CTSA MHC_I _T3 PTPN MHC_I _T3 PCK MHC_I _T3 IFNAR MHC_I _T3 IFNGR MHC_I _T3 TTC MHC_I _T3 MCM3AP MHC_I _T3 XRCC MHC_I _T3 SAMM MHC_I _T3 TOP2B MHC_I _T3 CTNNB MHC_I _T3 CDCP MHC_I _T3 SETD MHC_I _T3 PLXNB MHC_I _T3 SLC26A MHC_I _T3 TEX MHC_I _T3 PBRM MHC_I _T3 FLNB MHC_I _T3 UBA MHC_I _T3 CEP MHC_I _T3 HTT MHC_I _T3 RBM MHC_I _T3 OCIAD MHC_I _T3 SEC31A MHC_I _T3 OTUD MHC_I _T3 KLHL MHC_I _T3 BRD MHC_I _T3 KIAA MHC_I
55 _T3 ZFR MHC_I _T3 SKP MHC_I _T3 RASA MHC_I _T3 LNPEP MHC_I _T3 PPIP5K MHC_I _T3 TCERG MHC_I _T3 CLINT MHC_I _T3 LRRC16A MHC_I _T3 TAPBP MHC_I _T3 CUTA MHC_I _T3 FOXP MHC_I _T3 ORC MHC_I _T3 MDN MHC_I _T3 PTPRK MHC_I _T3 MED MHC_I _T3 INTS MHC_I _T3 HOXA MHC_I _T3 OGDH MHC_I _T3 GUSB MHC_I _T3 DTX MHC_I _T3 ANKIB MHC_I _T3 SLC25A MHC_I _T3 BAIAP2L MHC_I _T3 ZNHIT MHC_I _T3 COG MHC_I _T3 JHDM1D MHC_I _T3 BRAF MHC_I _T3 ZYX MHC_I _T3 R3HCC MHC_I _T3 ASPH MHC_I _T3 ARFGEF MHC_I
56 _T3 MAL MHC_I _T3 ATAD MHC_I _T3 EPPK MHC_I _T3 NPR MHC_I _T3 SLC35D MHC_I _T3 CIZ MHC_I _T3 ODF MHC_I _T3 TOR1A MHC_I _T3 ANAPC MHC_I X _T3 UBA MHC_I X _T3 FAAH MHC_I X _T3 SLC6A MHC_I FAP_G13 PRDM FAP_G13 USP FAP_G13 POGK FAP_G13 TRMT1L FAP_G13 SH3PXD2A FAP_G13 TCF7L FAP_G13 METTL FAP_G13 GTF2H FAP_G13 CSTF FAP_G13 ARHGAP FAP_G13 NOP FAP_G13 RECQL FAP_G13 RPAP FAP_G13 LRIG FAP_G13 USP FAP_G13 MIPEP FAP_G13 SUPT20H FAP_G13 DHRS4L FAP_G13 NYNRIN
57 FAP_G13 HECTD FAP_G13 HEATR5A FAP_G13 EFCAB FAP_G13 UBE3A FAP_G13 HERC FAP_G13 FANCI FAP_G13 CHD FAP_G13 COTL FAP_G13 NCOR FAP_G13 NCOR FAP_G13 TSEN FAP_G13 PIAS FAP_G13 BRD FAP_G13 RDH FAP_G13 ZNF FAP_G13 EIF2AK FAP_G13 BMPR FAP_G13 BMPR FAP_G13 BMPR FAP_G13 BMPR FAP_G13 ARFGAP FAP_G13 MINA FAP_G13 OPA FAP_G13 ATP13A FAP_G13 ATP8A FAP_G13 SEC24B FAP_G13 DDX FAP_G13 PRKAA FAP_G13 IL6ST FAP_G13 TMEM161B FAP_G13 BTNL
58 FAP_G13 AIM FAP_G13 CDC FAP_G13 PHF FAP_G13 VPS FAP_G13 FIGNL FAP_G13 STEAP FAP_G13 MEPCE FAP_G13 PNPLA FAP_G13 KAT6A FAP_G13 SDR16C FAP_G13 NBN FAP_G13 FZD FAP_G13 NFIL X FAP_G13 WDR X FAP_G13 TAZ FAP_G14 MOV FAP_G14 NR5A FAP_G14 PIK3C2B FAP_G14 AHCTF FAP_G14 CSTF2T FAP_G14 DNAJC FAP_G14 ZNF518A FAP_G14 BBIP FAP_G14 TCF7L FAP_G14 PLEKHA FAP_G14 PEX FAP_G14 SPI FAP_G14 CDC42BPG FAP_G14 DHCR FAP_G14 DLAT FAP_G14 LRP
59 FAP_G14 LRP FAP_G14 FBXO FAP_G14 BAZ1A FAP_G14 MAP4K FAP_G14 PTGR FAP_G14 UBE3A FAP_G14 TJP FAP_G14 SLC12A FAP_G14 MYO5C FAP_G14 HERC FAP_G14 MAP2K FAP_G14 EFTUD FAP_G14 NUBP FAP_G14 HEATR FAP_G14 MYO FAP_G14 CWC FAP_G14 XYLT FAP_G14 ANKRD FAP_G14 PPP4R FAP_G14 FUT FAP_G14 ATG4D FAP_G14 PSME FAP_G14 DUSP FAP_G14 HELZ FAP_G14 MINA FAP_G14 ZBTB FAP_G14 MYLK FAP_G14 ATP11B FAP_G14 OPA FAP_G14 IL6ST FAP_G14 TMEM161B
60 FAP_G14 FAM46A FAP_G14 FBXL FAP_G14 STEAP FAP_G14 MTERF FAP_G14 NBN FAP_G14 FAM83H FAP_G15 H6PD FAP_G15 USP FAP_G15 FAM63A FAP_G15 FBXW FAP_G15 SIRT FAP_G15 PDE3B FAP_G15 SORL FAP_G15 FBXL FAP_G15 PRIM FAP_G15 C12orf FAP_G15 UHRF1BP1L FAP_G15 GNPTAB FAP_G15 SUDS FAP_G15 HSPH FAP_G15 SLC7A FAP_G15 TC2N FAP_G15 PARN FAP_G15 NOMO FAP_G15 EARS FAP_G15 RSPRY FAP_G15 SMG FAP_G15 TMEM FAP_G15 ASNA FAP_G15 ZNF FAP_G15 ROCK
61 FAP_G15 PID FAP_G15 TRIP FAP_G15 ANO FAP_G15 ATXN FAP_G15 MINA FAP_G15 TOMM70A FAP_G15 ZXDC FAP_G15 NMNAT FAP_G15 PLOD FAP_G15 EHHADH FAP_G15 SEC24B FAP_G15 LARP1B FAP_G15 FHDC FAP_G15 LMBRD FAP_G15 ITGA FAP_G15 DHX FAP_G15 IL6ST FAP_G15 ANKRA FAP_G15 DAXX FAP_G15 DAXX FAP_G15 MED FAP_G15 PGM FAP_G15 AIM FAP_G15 CDC FAP_G15 REPS FAP_G15 KATNA FAP_G15 CASD FAP_G15 TRIM FAP_G15 GTF2E FAP_G15 WASH FAP_G15 CAAP
62 FAP_G15 NOL FAP_G15 EXD X FAP_G15 ABCB X FAP_G15 NDUFA X FAP_G15 AIFM FAP_G2 SMIM FAP_G2 MRPL FAP_G2 DEPDC FAP_G2 ZNF FAP_G2 NUF FAP_G2 NUP FAP_G2 AHCTF FAP_G2 KCNMA FAP_G2 CCSER FAP_G2 NSMCE4A FAP_G2 MED FAP_G2 TMEM194A FAP_G2 XPOT FAP_G2 NUP FAP_G2 UBE3B FAP_G2 CDK FAP_G2 HAUS FAP_G2 RALGAPA FAP_G2 BTBD FAP_G2 VPS13C FAP_G2 UBN FAP_G2 SMG FAP_G2 POLR3E FAP_G2 CTCF FAP_G2 RANBP FAP_G2 METTL
63 FAP_G2 SOX FAP_G2 DYM FAP_G2 KLK FAP_G2 REV FAP_G2 RIF FAP_G2 STAM FAP_G2 SATB FAP_G2 PCED1A FAP_G2 GPCPD FAP_G2 CLASP FAP_G2 CLASP FAP_G2 SEC22C FAP_G2 GBE FAP_G2 GPR FAP_G2 CWH FAP_G2 LARP1B FAP_G2 TTC FAP_G2 SHROOM FAP_G2 CSNK1A FAP_G2 HMMR FAP_G2 RPF FAP_G2 ARID1B FAP_G2 TRIM FAP_G2 LMBR FAP_G2 TACC FAP_G2 MTDH FAP_G2 LRRC FAP_G2 NOL X FAP_G2 TRMT2B X FAP_G2 ARMCX X FAP_G2 IDS
64 FAP_G25 RHBDL FAP_G25 RNF FAP_G25 ZNF FAP_G25 ACBD FAP_G25 SF FAP_G25 SLC11A FAP_G25 COPS FAP_G25 RAB27A FAP_G25 RPS17L FAP_G25 UNC45A FAP_G25 ATP6V0D FAP_G25 BAIAP FAP_G25 DSG FAP_G25 CBLC FAP_G25 ACTR FAP_G25 ORC FAP_G25 CD FAP_G25 GALNT FAP_G25 SLC41A FAP_G25 NCK FAP_G25 ETFDH FAP_G25 RIOK FAP_G25 SIL FAP_G25 PCDH FAP_G25 CCHCR FAP_G25 ATG FAP_G29 CLCA FAP_G29 WAPAL FAP_G29 FRA10AC FAP_G29 FGFR1OP FAP_G29 TC2N
65 FAP_G29 PDIA FAP_G29 GOSR FAP_G29 ZC3H FAP_G29 ATXN FAP_G29 NCK FAP_G29 TTC FAP_G29 SLC39A FAP_G29 ABCE FAP_G29 SKIV2L FAP_G29 HNRNPH FAP_G29 SENP X FAP_G29 BEX FAP_G3 ZMYM FAP_G3 ELOVL FAP_G3 FAF FAP_G3 UBE2Q FAP_G3 CEP FAP_G3 SSSCA FAP_G3 PPP6R FAP_G3 TEP FAP_G3 RALGAPA FAP_G3 PNMA FAP_G3 TRIP FAP_G3 BTBD FAP_G3 C15orf FAP_G3 SPPL2A FAP_G3 HERC FAP_G3 TRIP FAP_G3 NLRP FAP_G3 SENP FAP_G3 LAMA
66 FAP_G3 ZNF FAP_G3 ZNF FAP_G3 RDH FAP_G3 ANO FAP_G3 GRAMD1C FAP_G3 SLCO2A FAP_G3 ANKHD FAP_G3 ANKHD EIF4EBP FAP_G3 TBP FAP_G3 PPAPDC1B FAP_G3 PTPRD FAP_G3 RMI X FAP_G3 BTK FAP_G39 API FAP_G39 FIBP FAP_G39 ERGIC FAP_G39 SNX FAP_G39 BNIP FAP_G39 PDXDC FAP_G39 TCF FAP_G39 THRA FAP_G39 JUND FAP_G39 FRZB FAP_G39 HIBCH FAP_G39 FARSB FAP_G39 IGLL FAP_G39 RPP FAP_G39 MBD FAP_G39 OPA FAP_G39 PPP1R FAP_G39 CWC
67 FAP_G39 UBTD FAP_G39 ACAT FAP_G39 ADAMDEC X FAP_G39 NONO FAP_G47 EPB FAP_G47 EFCAB FAP_G47 MCOLN FAP_G47 FMO FAP_G47 PCGF FAP_G47 NEURL FAP_G47 ZFP FAP_G47 PCF FAP_G47 GOLT1B FAP_G47 PUS7L FAP_G47 CPSF FAP_G47 TBC1D FAP_G47 DLGAP FAP_G47 SIPA1L FAP_G47 BAG FAP_G47 CCNB FAP_G47 HEATR FAP_G47 CNOT FAP_G47 ATP6V0D FAP_G47 TEFM FAP_G47 THOC FAP_G47 ABHD FAP_G47 SLC39A FAP_G47 PRKD FAP_G47 PCBP FAP_G47 ZNF FAP_G47 MRPS
68 FAP_G47 ORC FAP_G47 PKP FAP_G47 GPCPD FAP_G47 C3orf FAP_G47 SKIL FAP_G47 NUP FAP_G47 CAMK2D FAP_G47 PLK FAP_G47 FBXO FAP_G47 SKIV2L FAP_G47 FEM1C FAP_G47 TMEM FAP_G47 TMEM FAP_G47 LMBR FAP_G47 FAM129B X FAP_G47 AIFM FAP_G48 MKNK FAP_G48 MKNK FAP_G48 FAF FAP_G48 EPC FAP_G48 PHLDA FAP_G48 ZFP FAP_G48 PCF FAP_G48 NXPE FAP_G48 LPAR FAP_G48 STYK FAP_G48 CPSF FAP_G48 APPL FAP_G48 CLIP FAP_G48 GOLGA FAP_G48 DTD
69 FAP_G48 ZNF FAP_G48 PGP FAP_G48 FOPNL FAP_G48 VPS FAP_G48 HEATR FAP_G48 CNOT FAP_G48 ATP6V0D FAP_G48 ESRP FAP_G48 TOP2A FAP_G48 ABCA FAP_G48 THOC FAP_G48 PIGN FAP_G48 ZBTB7A FAP_G48 ZNF FAP_G48 PEPD FAP_G48 PUM FAP_G48 CALM FAP_G48 MOGS FAP_G48 EIF5B FAP_G48 ORC FAP_G48 KIAA FAP_G48 GPCPD FAP_G48 PAXBP FAP_G48 GART FAP_G48 EIF4E FAP_G48 GATAD FAP_G48 WNK FAP_G5 EIF4G FAP_G5 KIAA0319L FAP_G5 NRD FAP_G5 UBAP2L
70 FAP_G5 ELF FAP_G5 FAM21B FAP_G5 USP FAP_G5 TTC FAP_G5 PRPF FAP_G5 IPO FAP_G5 SMARCD FAP_G5 USP FAP_G5 GNS FAP_G5 MTERFD FAP_G5 ANKRD13A FAP_G5 DDX FAP_G5 ARHGAP FAP_G5 LRR FAP_G5 FBXO FAP_G5 MTMR FAP_G5 RMDN FAP_G5 KIAA FAP_G5 CLCN FAP_G5 PSKH FAP_G5 DHX FAP_G5 ARHGAP FAP_G5 MYO18A FAP_G5 EZH FAP_G5 CEP FAP_G5 ROCK FAP_G5 ZADH FAP_G5 PTBP FAP_G5 DNMT FAP_G5 ROCK FAP_G5 SOS
71 FAP_G5 PSME FAP_G5 USP FAP_G5 POLR1B FAP_G5 CDCA FAP_G5 HIBCH FAP_G5 SP FAP_G5 PLCB FAP_G5 PAXBP FAP_G5 THUMPD FAP_G5 OXSR FAP_G5 SMARCC FAP_G5 CCDC FAP_G5 FAM208A FAP_G5 ZBTB FAP_G5 CD FAP_G5 PARP FAP_G5 SMC FAP_G5 MCCC FAP_G5 EXOC FAP_G5 TIGD FAP_G5 PPP3CA FAP_G5 TBCK FAP_G5 FNIP FAP_G5 ELOVL FAP_G5 RAD FAP_G5 LRRC16A FAP_G5 PAQR FAP_G5 SESN FAP_G5 TRIM FAP_G5 TMEM FAP_G5 RBM
72 FAP_G5 NBN FAP_G5 ERMP X FAP_G5 THOC FAP_G53 SNRNP FAP_G53 ZNF FAP_G53 SUCO FAP_G53 MIA FAP_G53 FAM171A FAP_G53 ARHGAP FAP_G53 ZNF33A FAP_G53 GPRIN FAP_G53 WAPAL FAP_G53 METTL FAP_G53 NUP FAP_G53 RSF FAP_G53 ERGIC FAP_G53 IPO FAP_G53 R3HDM FAP_G53 MIS18BP FAP_G53 UBE3A FAP_G53 DUOXA FAP_G53 MYO1E FAP_G53 CLPX FAP_G53 ZC3H7A FAP_G53 GLG FAP_G53 MIS FAP_G53 MIS FAP_G53 MED FAP_G53 EMC FAP_G53 PPP1R12C FAP_G53 EIF2AK
73 FAP_G53 RBM FAP_G53 STK FAP_G53 SBF FAP_G53 DALRD FAP_G53 ABHD FAP_G53 SLC41A FAP_G53 ABTB FAP_G53 FAM175A FAP_G53 ABCE FAP_G53 YIPF FAP_G53 THG1L FAP_G53 KIF13A FAP_G53 PEX FAP_G53 SLC17A FAP_G53 BCKDHB FAP_G53 TMEM184A FAP_G53 ZHX FAP_G53 FBXW FAP_G55 WDTC FAP_G55 PDE4DIP FAP_G55 TTC FAP_G55 ARHGAP FAP_G55 C11orf FAP_G55 KDM5A FAP_G55 MED FAP_G55 ERGIC FAP_G55 XPOT FAP_G55 CAND FAP_G55 SART FAP_G55 RALGAPA FAP_G55 STAT
74 FAP_G55 PSME FAP_G55 ZMYND FAP_G55 TOB FAP_G55 ATP13A FAP_G55 FNIP FAP_G55 C5orf FAP_G55 SLC17A FAP_G55 MLLT FAP_G55 SAMD FAP_G55 TRIM FAP_G55 PNPLA FAP_G55 IARS X FAP_G55 UPF3B FAP_G6 EXOSC FAP_G6 NRD FAP_G6 SRSF FAP_G6 ODF2L FAP_G6 NBPF FAP_G6 DDX FAP_G6 USP FAP_G6 R3HCC1L FAP_G6 ZBED FAP_G6 TTC FAP_G6 CDC42BPG FAP_G6 PPFIA FAP_G6 SLC11A FAP_G6 USP FAP_G6 XPOT FAP_G6 ATP2B FAP_G6 ANKRD13A FAP_G6 MBNL
75 FAP_G6 SECISBP2L FAP_G6 ZC3H7A FAP_G6 RRN FAP_G6 POLR3E FAP_G6 DHX FAP_G6 MYO18A FAP_G6 ROCK FAP_G6 STXBP FAP_G6 ZNF FAP_G6 FCGBP FAP_G6 ROCK FAP_G6 PRKD FAP_G6 SOS FAP_G6 PTCD FAP_G6 EIF2AK FAP_G6 GALNT FAP_G6 FERMT FAP_G6 TTLL FAP_G6 TOP2B FAP_G6 OXSR FAP_G6 STAG FAP_G6 UGT2A FAP_G6 FRG FAP_G6 SLC30A FAP_G6 SLC12A FAP_G6 RANBP FAP_G6 MARCKS FAP_G6 AKAP FAP_G6 CFTR FAP_G6 ERMP FAP_G6 KDM4C
76 FAP_G6 TOPORS FAP_G6 SMC FAP_G6 TMEM X FAP_G6 ZBED X FAP_G6 THOC FAP_G61 PQLC FAP_G61 TRIT FAP_G61 OMA FAP_G61 YME1L FAP_G61 SMNDC FAP_G61 TCF7L FAP_G61 PPFIA FAP_G61 CCDC FAP_G61 LRIG FAP_G61 MED13L FAP_G61 NHLRC FAP_G61 COG FAP_G61 LPAR FAP_G61 ARID4A FAP_G61 DICER FAP_G61 DYNC1H FAP_G61 BNIP FAP_G61 NEURL FAP_G61 CDK FAP_G61 SPOP FAP_G61 TRIM FAP_G61 AXIN FAP_G61 MOB3A FAP_G61 SH3GL FAP_G61 DPP FAP_G61 FCGBP
77 FAP_G61 NOL FAP_G61 RANBP FAP_G61 BAZ2B FAP_G61 PLAGL FAP_G61 NCOA FAP_G61 YTHDF FAP_G61 SCAF FAP_G61 SCAF FAP_G61 TCAIM FAP_G61 APEH FAP_G61 TWF FAP_G61 ZDHHC FAP_G61 TOPBP FAP_G61 CEP FAP_G61 ATR FAP_G61 PLOD FAP_G61 FRYL FAP_G61 SNX FAP_G61 KDM3B FAP_G61 RGL FAP_G61 ASCC FAP_G61 MICAL FAP_G61 RFC FAP_G61 TRIM FAP_G61 NUP FAP_G61 TMUB FAP_G61 TRPM X FAP_G61 HUWE FAP_G67 SCMH FAP_G67 FMO FAP_G67 CEP
78 FAP_G67 ZDHHC FAP_G67 MANSC FAP_G67 FKBP FAP_G67 TBX FAP_G67 PYGL FAP_G67 GNPNAT FAP_G67 TRIM FAP_G67 LMNB FAP_G67 ORC FAP_G67 SF3B FAP_G67 MED FAP_G67 SACM1L FAP_G67 PCBP FAP_G67 UBA FAP_G67 AP3B FAP_G67 CDC FAP_G67 AHCYL FAP_G67 TLE X FAP_G67 UBE2A _EBL1 ZBTB _EBL1 PDSS _EBL1 PBLD _EBL1 ACY _EBL1 LPCAT _EBL1 AQR _EBL1 VWA _EBL1 GEMIN _EBL1 VAT _EBL1 NPEPPS _EBL1 NPEPPS _EBL1 HOXB
79 _EBL1 ATP13A _EBL1 ZNF _EBL1 DUSP _EBL1 HGD _EBL1 PARP _EBL1 SEC31A _EBL1 FAF _EBL1 SESN _EBL1 INTS _EBL1 SECISBP X _EBL1 ARSE X _EBL1 PSMD _EBL11 GPR _EBL11 PLEKHM _EBL11 EPHA _EBL11 HSPG _EBL11 STX _EBL11 EPB _EBL11 TINAGL _EBL11 YARS _EBL11 ZMYM _EBL11 PABPC _EBL11 PLK _EBL11 PLK _EBL11 HSPB _EBL11 CYP2J _EBL11 LRRC8D _EBL11 TRIM _EBL11 POGZ _EBL11 DENND4B _EBL11 THBS
80 _EBL11 CLK _EBL11 DEDD _EBL11 B4GALT _EBL11 C4BPB _EBL11 LBR _EBL11 ARHGAP _EBL11 PRTFDC _EBL11 ZNF33B _EBL11 MMS _EBL11 PYROXD _EBL11 DMBT _EBL11 ATHL _EBL11 SIGIRR _EBL11 PNPLA _EBL11 TIMM10B _EBL11 ARHGAP _EBL11 FNBP _EBL11 NUP _EBL11 VPS37C _EBL11 INTS _EBL11 ARL _EBL11 RIN _EBL11 CTSF _EBL11 CORO1B _EBL11 PPP6R _EBL11 NUMA _EBL11 PGM2L _EBL11 PRKRIR _EBL11 PRSS _EBL11 HYOU _EBL11 EI _EBL11 LPAR
81 _EBL11 CTN _EBL11 SCAF _EBL11 MYO1A _EBL11 XPOT _EBL11 TDG _EBL11 TBX _EBL11 WSB _EBL11 TAOK _EBL11 CLIP _EBL11 NCOR _EBL11 FBXL _EBL11 MCF2L _EBL11 CHD _EBL11 SLC7A _EBL11 PTPN _EBL11 ANKRD _EBL11 CDC42BPB _EBL11 PACS _EBL11 SMAD _EBL11 SCAMP _EBL11 ZNF _EBL11 MAPK8IP _EBL11 KIAA _EBL11 ZNF _EBL11 PHKG _EBL11 LPCAT _EBL11 NLRC _EBL11 PLEKHG _EBL11 FUK _EBL11 GLG _EBL11 MLKL
82 _EBL11 PIEZO _EBL11 DEF _EBL11 GEMIN _EBL11 FLII _EBL11 GIT _EBL11 PSMD _EBL11 HNF1B _EBL11 KAT2A _EBL11 STAT5A _EBL11 TUBG _EBL11 PLCD _EBL11 SPOP _EBL11 HGS _EBL11 C17orf _EBL11 FOXK _EBL11 PPP4R _EBL11 NAPG _EBL11 THOP _EBL11 FUT _EBL11 CAMSAP _EBL11 C19orf _EBL11 DNMT _EBL11 KEAP _EBL11 SMARCA _EBL11 CC2D1A _EBL11 AKAP8L _EBL11 CHERP _EBL11 MYO9B _EBL11 PIK3R _EBL11 ZNF _EBL11 SARS
83 _EBL11 DMPK _EBL11 PRKD _EBL11 CCDC _EBL11 ZNF _EBL11 ZNF _EBL11 ZNF _EBL11 ADAM _EBL11 ASXL _EBL11 ZNF _EBL11 BIRC _EBL11 TTC _EBL11 RMND5A _EBL11 THNSL _EBL11 SNRNP _EBL11 SLC9A _EBL11 MYO7B _EBL11 GALNT _EBL11 ACVR _EBL11 PKP _EBL11 KIAA _EBL11 G _EBL11 AGAP _EBL11 CEBPB _EBL11 PCMTD _EBL11 DOPEY _EBL11 AGPAT _EBL11 PI4KA _EBL11 MAPK _EBL11 BCR _EBL11 SMARCB _EBL11 SF3A
84 _EBL11 PRR14L _EBL11 IFT _EBL11 MPST _EBL11 CSNK1E _EBL11 ZBED _EBL11 TUBGCP _EBL11 PLXNB _EBL11 BHLHE _EBL11 CAMK _EBL11 TAMM _EBL11 TRANK _EBL11 SEC22C _EBL11 PTPN _EBL11 PRKCD _EBL11 GPR _EBL11 MCM _EBL11 ABTB _EBL11 EIF4A _EBL11 NELFA _EBL11 MAN2B _EBL11 SRD5A _EBL11 DCK _EBL11 COQ _EBL11 UGT _EBL11 TMEM184C _EBL11 TMEM _EBL11 CLCN _EBL11 CLCN _EBL11 MATR _EBL11 PCDH _EBL11 LARP
85 _EBL11 GRK _EBL11 MAML _EBL11 ECI _EBL11 RREB _EBL11 HIST1H2BK _EBL11 VARS _EBL11 MICA _EBL11 BAG _EBL11 RXRB _EBL11 UBR _EBL11 TBCC _EBL11 TUBE _EBL11 FRK _EBL11 NCOA _EBL11 EPB41L _EBL11 NHSL _EBL11 PHF _EBL11 NFE2L _EBL11 TAX1BP _EBL11 TRRAP _EBL11 SND _EBL11 LUZP _EBL11 ARHGEF _EBL11 AGAP _EBL11 AGAP _EBL11 HR _EBL11 KIAA _EBL11 TCEA _EBL11 SLC25A _EBL11 RNF _EBL11 MTSS
86 _EBL11 TOP1MT _EBL11 EEF1D _EBL11 FAM83H _EBL11 SLC52A _EBL11 CPSF _EBL11 KIAA _EBL11 CAAP _EBL11 UBAP _EBL11 RGP _EBL11 TMEM8B _EBL11 GNE _EBL11 ZBTB _EBL11 GCNT _EBL11 NCBP _EBL11 PTPN _EBL11 NTMT _EBL11 ASB _EBL11 TSC _EBL11 TSC _EBL11 NOXA X _EBL11 MED X _EBL11 CASK X _EBL11 HUWE X _EBL11 HDAC X _EBL11 AIFM _EBL13 SFPQ _EBL13 UCHL _EBL13 EPC _EBL13 MICU _EBL13 FNBP _EBL13 ZDHHC
87 _EBL13 PEX _EBL13 PAN _EBL13 TC2N _EBL13 AMN _EBL13 NDN _EBL13 AMDHD _EBL13 AXIN _EBL13 ZBTB7A _EBL13 SIRT _EBL13 FUT _EBL13 TYK _EBL13 DDA _EBL13 ANKRD _EBL13 VRK _EBL13 PPP6R _EBL13 ZNF _EBL13 MAPRE _EBL13 ZFP36L _EBL13 AP1B _EBL13 APOL _EBL13 SBF _EBL13 CPT1B _EBL13 UBP _EBL13 NICN _EBL13 TPRA _EBL13 ABCG _EBL13 ZCCHC _EBL13 SHROOM _EBL13 ARHGAP _EBL13 GIMAP _EBL13 TMEM
88 _EBL13 C9orf X _EBL13 G6PD _EBL18 SMPD _EBL18 SVIP _EBL18 CPT1A _EBL18 TM9SF _EBL18 BMP _EBL18 ATP6V0D _EBL18 RBKS _EBL18 MEMO _EBL18 HGD _EBL18 KLF _EBL20 VANGL _EBL20 RPRD _EBL20 ZNF _EBL20 SLC25A _EBL20 ASPM _EBL20 CENPF _EBL20 SIRT _EBL20 RRP _EBL20 SCD _EBL20 MOB _EBL20 KBTBD _EBL20 MAP3K _EBL20 ACY _EBL20 ARAP _EBL20 ZW _EBL20 ETS _EBL20 NOP _EBL20 CTN _EBL20 ADCY
89 _EBL20 SLC11A _EBL20 LRP _EBL20 NTN _EBL20 OAS _EBL20 MTMR _EBL20 TUBGCP _EBL20 TUBGCP _EBL20 ARHGAP _EBL20 DCAF _EBL20 ELMSAN _EBL20 SPATA _EBL20 AQR _EBL20 TBC1D2B _EBL20 GFER _EBL20 MT _EBL20 PDXDC _EBL20 RRN _EBL20 LYRM _EBL20 ZG _EBL20 ZNF _EBL20 GPR _EBL20 ZCCHC _EBL20 CENPV _EBL20 DHX _EBL20 TLK _EBL20 PSMC _EBL20 HID _EBL20 LLGL _EBL20 RNF _EBL20 FASN _EBL20 CHAF1A
90 _EBL20 TICAM _EBL20 MLLT _EBL20 RAVER _EBL20 DYRK1B _EBL20 PVRL _EBL20 LIG _EBL20 POLD _EBL20 ZNF _EBL20 DDX _EBL20 NCOA _EBL20 ADCY _EBL20 PNPT _EBL20 USP _EBL20 NCK _EBL20 SAP _EBL20 NDUFS _EBL20 SGPP _EBL20 MFF _EBL20 FASTKD _EBL20 CENPB _EBL20 C20orf _EBL20 FAM83D _EBL20 ATP9A _EBL20 HELZ _EBL20 UCKL _EBL20 PI4KA _EBL20 ACO _EBL20 XPC _EBL20 HYAL _EBL20 PROS _EBL20 ZBTB
91 _EBL20 RPN _EBL20 RNF _EBL20 NELFA _EBL20 NIPAL _EBL20 FRYL _EBL20 HERC _EBL20 SEC24D _EBL20 KIAA _EBL20 PPID _EBL20 ING _EBL20 TRAPPC _EBL20 AP3B _EBL20 PCSK _EBL20 SRP _EBL20 PGGT1B _EBL20 SEMA6A _EBL20 CSNK1G _EBL20 TMEM63B _EBL20 NHSL _EBL20 CCZ1B _EBL20 ANLN _EBL20 IMMP2L _EBL20 MEST _EBL20 XPO _EBL20 PDLIM _EBL20 INTS _EBL20 DDHD _EBL20 WHSC1L _EBL20 NBN _EBL20 CPSF _EBL20 ZNF
92 _EBL20 NOL _EBL20 ZCCHC _EBL20 SECISBP _EBL20 TGFBR _EBL20 RNF _EBL20 NSMF X _EBL20 TRMT2B X _EBL20 TRMT2B _EBL23 AMPD _EBL23 FGFR _EBL23 NSMCE4A _EBL23 BATF _EBL23 TBRG _EBL23 WDR _EBL23 STAT _EBL23 RNFT _EBL23 JOSD _EBL23 WDR _EBL23 TTC _EBL23 MST1R _EBL23 ABCG _EBL23 GALNT _EBL23 TRIM _EBL26 FRRS _EBL26 KIFAP _EBL26 PLXNA _EBL26 IPMK _EBL26 JMJD1C _EBL26 TUBGCP _EBL26 SBF _EBL26 KLRB
93 _EBL26 RPAP _EBL26 TBX _EBL26 EGLN _EBL26 AMFR _EBL26 ATP6V0D _EBL26 ZBTB _EBL26 ATP13A _EBL26 ATP13A _EBL26 STRN _EBL26 ATG9A _EBL26 CRNKL _EBL26 PIGP _EBL26 CHEK _EBL26 TRANK _EBL26 PBRM _EBL26 FAM208A _EBL26 DNAJC _EBL26 TFDP _EBL26 PLD _EBL26 FXR _EBL26 MCCC _EBL26 ATP8A _EBL26 TTC _EBL26 CUL _EBL26 ANKMY _EBL26 KIAA X _EBL26 ARHGEF X _EBL26 DIAPH X _EBL26 TMLHE _EBL3 KDM4A _EBL3 FAM171A
94 _EBL3 ZMIZ _EBL3 ATN _EBL3 PTPRR _EBL3 MTERFD _EBL3 CPSF _EBL3 ATXN2L _EBL3 BBS _EBL3 ATP6V0D _EBL3 ADORA2B _EBL3 ERAL _EBL3 LIPG _EBL3 MED _EBL3 ARHGEF _EBL3 ZNF _EBL3 LMAN2L _EBL3 TOPBP _EBL3 SH3D _EBL3 RWDD _EBL3 FRG _EBL3 SKP _EBL3 YIPF _EBL3 VARS _EBL3 MED _EBL3 PCMTD _EBL3 ARFGEF _EBL3 INIP X _EBL3 MTM _EBL5 DHX _EBL5 1-Mar _EBL5 NT5C _EBL5 USP
95 _EBL5 TBL _EBL5 CWC _EBL5 MYO5B _EBL5 SERTAD _EBL5 CBWD _EBL5 PPIL _EBL5 PLOD _EBL5 LRRC _EBL5 TBC1D _EBL5 CORO2A _EBL7 EIF4G _EBL7 FOXJ _EBL7 POLL _EBL7 TNKS1BP _EBL7 EXPH _EBL7 RECQL _EBL7 ZFC3H _EBL7 CRY _EBL7 OAS _EBL7 ATP6V0A _EBL7 BTBD _EBL7 AQR _EBL7 FAM214A _EBL7 PEAK _EBL7 KIAA _EBL7 PLCD _EBL7 COIL _EBL7 MFSD _EBL7 TBCD _EBL7 PTPN _EBL7 ZBTB7A
96 _EBL7 KEAP _EBL7 MTA _EBL7 RNF _EBL7 KIAA _EBL7 KIF3B _EBL7 DLG _EBL7 HERC _EBL7 TLR _EBL7 FAT _EBL7 CCDC _EBL7 MYO _EBL7 ZCCHC _EBL7 FAM8A _EBL7 C6orf _EBL7 C6orf _EBL7 FBXL _EBL7 CCDC28A _EBL7 INTS _EBL7 FTSJ _EBL7 PEX _EBL7 WWP _EBL7 FBXW _EBL7 SETX X _EBL7 ELF _EBL9 AGRN _EBL9 ACAP _EBL9 DVL _EBL9 ATAD3A _EBL9 CDK11B _EBL9 SKI _EBL9 MTHFR
97 _EBL9 MTHFR _EBL9 VPS13D _EBL9 PLEKHM _EBL9 EPHA _EBL9 EPHA _EBL9 SDHB _EBL9 HP1BP _EBL9 USP _EBL9 USP _EBL9 USP _EBL9 HSPG _EBL9 SRSF _EBL9 WASF _EBL9 YARS _EBL9 RNF19B _EBL9 IPO _EBL9 MAST _EBL9 CDKN2C _EBL9 INADL _EBL9 DNAJB _EBL9 HENMT _EBL9 PTGFRN _EBL9 PRKAB _EBL9 VPS _EBL9 THEM _EBL9 NDUFS _EBL9 POGK _EBL9 XPR _EBL9 PLXNA _EBL9 SLC30A _EBL9 EPHX
98 _EBL9 RBM _EBL9 NID _EBL9 NID _EBL9 ARHGAP _EBL9 ARHGAP _EBL9 DNAJC _EBL9 SORBS _EBL9 ZNF518A _EBL9 NFKB _EBL9 SMC _EBL9 SHOC _EBL9 DCLRE1A _EBL9 SFXN _EBL9 MCMBP _EBL9 MKI _EBL9 MKI _EBL9 EPS8L _EBL9 ZNF _EBL9 SBF _EBL9 CKAP _EBL9 SLC43A _EBL9 DDB _EBL9 MYRF _EBL9 HNRNPUL _EBL9 MARK _EBL9 PLCB _EBL9 CDC42BPG _EBL9 EHD _EBL9 VPS _EBL9 SIPA _EBL9 SF3B
99 _EBL9 PITPNM _EBL9 SESN _EBL9 BIRC _EBL9 SIK _EBL9 NXPE _EBL9 HYOU _EBL9 DPAGT _EBL9 HINFP _EBL9 ARHGAP _EBL9 KDM5A _EBL9 KDM5A _EBL9 TNFRSF1A _EBL9 DUSP _EBL9 ITPR _EBL9 FAM60A _EBL9 PKP _EBL9 ACVRL _EBL9 SPRYD _EBL9 DGKA _EBL9 TIMELESS _EBL9 METTL _EBL9 MON _EBL9 TMEM _EBL9 CPM _EBL9 GLIPR _EBL9 GCN1L _EBL9 VPS33A _EBL9 SBNO _EBL9 RNF _EBL9 CDX _EBL9 FOXO
100 _EBL9 VWA _EBL9 EPSTI _EBL9 FBXL _EBL9 RBM _EBL9 TEP _EBL9 ACIN _EBL9 DHRS _EBL9 NYNRIN _EBL9 NYNRIN _EBL9 MIS18BP _EBL9 NIN _EBL9 FBXO _EBL9 PRKCH _EBL9 ACTN _EBL9 YLPM _EBL9 ACYP _EBL9 CCDC88C _EBL9 DICER _EBL9 ZNF _EBL9 BUB1B _EBL9 SPG _EBL9 SECISBP2L _EBL9 MYO5C _EBL9 BNIP _EBL9 HERC _EBL9 KIAA _EBL9 UACA _EBL9 TMEM8A _EBL9 RAB11FIP _EBL9 WDR _EBL9 HAGH
101 _EBL9 TSC _EBL9 PKD _EBL9 CREBBP _EBL9 UBALD _EBL9 TXNDC _EBL9 RBBP _EBL9 MVP _EBL9 E2F _EBL9 COG _EBL9 ACSF _EBL9 TUBB _EBL9 ATP2A _EBL9 ZZEF _EBL9 ZZEF _EBL9 SPNS _EBL9 MINK _EBL9 NUP _EBL9 TNK _EBL9 MD _EBL9 CHD _EBL9 C17orf _EBL9 SREBF _EBL9 NLK _EBL9 EVI2B _EBL9 ZNF _EBL9 MYO _EBL9 ERBB _EBL9 NT5C3B _EBL9 DHX _EBL9 WNK _EBL9 HDAC
102 _EBL9 PLEKHM _EBL9 RNFT _EBL9 APPBP _EBL9 TMC _EBL9 RAC _EBL9 FASN _EBL9 USP _EBL9 EPB41L _EBL9 TUBB _EBL9 ROCK _EBL9 TRAPPC _EBL9 POLRMT _EBL9 WDR _EBL9 MIDN _EBL9 REXO _EBL9 CSNK1G _EBL9 STAP _EBL9 TRAPPC _EBL9 ZNF _EBL9 C19orf _EBL9 RAVER _EBL9 ATG4D _EBL9 SMARCA _EBL9 CALR _EBL9 TRMT _EBL9 NACC _EBL9 PKN _EBL9 ZNF _EBL9 ANKRD _EBL9 ANKRD _EBL9 ZNF
103 _EBL9 USF _EBL9 FCGBP _EBL9 CLPTM _EBL9 FOXA _EBL9 DBP _EBL9 NOSIP _EBL9 SCAF _EBL9 IRF _EBL9 ZNF _EBL9 PRPF _EBL9 LENG _EBL9 HSPBP _EBL9 ZNF _EBL9 NOL _EBL9 NBAS _EBL9 ZFP36L _EBL9 RHOQ _EBL9 MSH _EBL9 PSME _EBL9 MTIF _EBL9 PNPT _EBL9 USP _EBL9 GCFC _EBL9 PROM _EBL9 SNRNP _EBL9 TMEM87B _EBL9 TMEM185B _EBL9 TMEM185B _EBL9 PKP _EBL9 LY _EBL9 PPIG
104 _EBL9 TRIP _EBL9 SH3BP _EBL9 PET _EBL9 RALGAPA _EBL9 KIF3B _EBL9 NECAB _EBL9 TRPC4AP _EBL9 PHF _EBL9 PKIG _EBL9 ADNP _EBL9 HELZ _EBL9 PAXBP _EBL9 GART _EBL9 DOPEY _EBL9 MX _EBL9 ABCG _EBL9 AIFM _EBL9 YDJC _EBL9 PPM1F _EBL9 BCR _EBL9 MYH _EBL9 DDX _EBL9 BRD _EBL9 TUBGCP _EBL9 PLXNB _EBL9 ARSA _EBL9 FANCD _EBL9 NUP _EBL9 CDCP _EBL9 RBM _EBL9 FLNB
105 _EBL9 TMF _EBL9 GBE _EBL9 CPOX _EBL9 IFT _EBL9 PVRL _EBL9 SPICE _EBL9 GOLGB _EBL9 EEFSEC _EBL9 MRPL _EBL9 HLTF _EBL9 LRRC _EBL9 FNDC3B _EBL9 DVL _EBL9 EPHB _EBL9 DGKQ _EBL9 DGKQ _EBL9 MFSD _EBL9 FRYL _EBL9 BMP2K _EBL9 ALPK _EBL9 EXOSC _EBL9 OTUD _EBL9 NR3C _EBL9 CTSO _EBL9 FAT _EBL9 PDCD _EBL9 6-Mar _EBL9 TRIO _EBL9 ZNF _EBL9 TARS _EBL9 NIPBL
106 _EBL9 ELOVL _EBL9 MRPS _EBL9 ANKRA _EBL9 GFM _EBL9 COL4A3BP _EBL9 WDR _EBL9 DHFR _EBL9 ELL _EBL9 ELL _EBL9 RIOK _EBL9 CDC42SE _EBL9 DIAPH _EBL9 CYFIP _EBL9 CCNG _EBL9 DDX _EBL9 RMND5B _EBL9 TRIM _EBL9 NFKBIL _EBL9 PRRC2A _EBL9 EHMT _EBL9 NELFE _EBL9 C6orf _EBL9 KCNK _EBL9 GLTSCR1L _EBL9 PEX _EBL9 PEX _EBL9 MRPS18A _EBL9 CDC5L _EBL9 FBXL _EBL9 TSPYL _EBL9 RMND
107 _EBL9 MTRF1L _EBL9 SNX _EBL9 SNX _EBL9 DPY19L _EBL9 DPY19L _EBL9 POLM _EBL9 TBRG _EBL9 DDC _EBL9 COBL _EBL9 BAZ1B _EBL9 DTX _EBL9 SAMD9L _EBL9 CCDC _EBL9 ACHE _EBL9 MUC _EBL9 LRWD _EBL9 IMMP2L _EBL9 POT _EBL9 CASP _EBL9 ABCB _EBL9 DNAJB _EBL9 SLC39A _EBL9 GTF2E _EBL9 KAT6A _EBL9 NSMAF _EBL9 CHD _EBL9 ASPH _EBL9 ARFGEF _EBL9 STAU _EBL9 IMPA _EBL9 TATDN
108 _EBL9 ST3GAL _EBL9 PTK _EBL9 OPLAH _EBL9 CPSF _EBL9 CBWD _EBL9 KIAA _EBL9 KIAA _EBL9 DENND4C _EBL9 MLLT _EBL9 TMEM _EBL9 TRPM _EBL9 IARS _EBL9 XPA _EBL9 GALNT _EBL9 KLF _EBL9 SLC31A _EBL9 TRIM _EBL9 STOM _EBL9 STRBP _EBL9 SLC25A _EBL9 DNM _EBL9 NUP _EBL9 NUP _EBL9 RAPGEF _EBL9 SETX X _EBL9 PRKX X _EBL9 STS X _EBL9 BCOR X _EBL9 BCOR X _EBL9 PHF X _EBL9 ATRX
109 X _EBL9 HNRNPH X _EBL9 PRPS X _EBL9 P X _EBL9 MBNL X _EBL9 SLC6A X _EBL9 BCAP _T1 MTOR _T1 MACF _T1 POMGNT _T1 SMG _T1 HSD17B _T1 EPRS _T1 PTEN _T1 PTEN _T1 ILK _T1 SBF _T1 TTC9C _T1 SCAF _T1 ZFC3H _T1 APAF _T1 ANAPC _T1 POLE _T1 TPP _T1 TPP _T1 TFDP _T1 SYNE _T1 ZFP36L _T1 AHSA _T1 DYNC1H _T1 BOLA _T1 STX
110 _T1 RBL _T1 DDX19A _T1 RFWD _T1 SPG _T1 TUBB _T1 ARRB _T1 NCOR _T1 SPAG _T1 RNF _T1 MOCOS _T1 PLD _T1 TOMM _T1 BIRC _T1 EPAS _T1 EIF5B _T1 ERCC _T1 NOSTRIN _T1 NOP _T1 SON _T1 CABIN _T1 TCF _T1 TADA _T1 NBEAL _T1 UBA _T1 PVRL _T1 PARP _T1 PLOD _T1 FNDC3B _T1 ZNF _T1 SNX _T1 PAM
111 _T1 IRF _T1 NDFIP _T1 TBC1D _T1 HLA-A _T1 ITPR _T1 EPB41L _T1 PEX _T1 MTHFD1L _T1 DDC _T1 EXT _T1 TAF _T1 TRAPPC _T1 EPPK _T1 TLN _T1 SMC _T2 EPHA _T2 C1QA _T2 SNRNP _T2 HDAC _T2 JUN _T2 GBP _T2 TARS _T2 PMVK _T2 TPR _T2 HNRNPU _T2 PTEN _T2 MUC5B _T2 FAR _T2 CAPRIN _T2 ST _T2 USP
112 _T2 PLBD _T2 DDX _T2 DDX _T2 LRP _T2 USP _T2 APAF _T2 ERP _T2 CHD _T2 CHD _T2 MAP4K _T2 SIPA1L _T2 DT _T2 SMEK _T2 VRK _T2 CDC42BPB _T2 HERC _T2 HERC _T2 TMEM87A _T2 UACA _T2 NEO _T2 GSPT _T2 SMG _T2 XPO _T2 ATXN2L _T2 PHKB _T2 ATP6V0D _T2 CIRH1A _T2 ANKRD _T2 ACADVL _T2 SREBF _T2 FLII
113 _T2 PRPSAP _T2 ALDOC _T2 CPD _T2 UTP _T2 GSDMB _T2 ATP6V0A _T2 GRN _T2 KPNB _T2 PSMD _T2 BPTF _T2 SRP _T2 DSC _T2 DYM _T2 AP3D _T2 HDGFRP _T2 NDUFA _T2 ASNA _T2 BABAM _T2 SPINT _T2 SUPT5H _T2 AKT _T2 SAE _T2 MSH _T2 MSH _T2 SNRNP _T2 MMADHC _T2 NDUFS _T2 ACSL _T2 HM _T2 SAMHD _T2 PARVB
114 _T2 MLH _T2 WDR _T2 WDR _T2 MUC _T2 RNF _T2 TMEM _T2 G3BP _T2 SEC24D _T2 SMARCA _T2 LRBA _T2 FAT _T2 TRIO _T2 MYO _T2 NIPBL _T2 ITGA _T2 MCCC _T2 HMGCR _T2 HIST1H1B _T2 DCBLD _T2 HBS1L _T2 TRRAP _T2 CYP3A _T2 MUC _T2 MUC _T2 MCM _T2 KIAA _T2 DCTN _T2 VPS13A _T2 NANS _T2 GLE _T2 PMPCA
115 _T2 NOTCH _T2 TUBB4B X _T2 ACOT X _T2 USP9X X _T2 MAGT _T3 CPSF3L _T3 DVL _T3 RERE _T3 RERE _T3 CTN _T3 KIF1B _T3 VPS13D _T3 PRDM _T3 NECAP _T3 NECAP _T3 UBR _T3 EMC _T3 USP _T3 C1orf _T3 RNF19B _T3 SFPQ _T3 PTPRF _T3 ZFYVE _T3 PKN _T3 DPYD _T3 RFX _T3 TUFT _T3 C1orf _T3 APOA1BP _T3 ADIPOR _T3 NEK
116 _T3 LBR _T3 HIST3H2A _T3 CEP _T3 GTPBP _T3 ACBD _T3 FAM21B _T3 DDX _T3 USP _T3 CEP _T3 ALDH18A _T3 GBF _T3 C10orf _T3 FAM45A _T3 WDR _T3 BUB _T3 ANO _T3 MUC5B _T3 MUC5B _T3 PIK3C2A _T3 ARL14EP _T3 CTNND _T3 SF _T3 VPS _T3 VPS _T3 SYVN _T3 SCYL _T3 DPP _T3 RPS6KB _T3 RSF _T3 SYTL _T3 KIAA
117 _T3 CASP _T3 SIDT _T3 SRPR _T3 ARHGAP _T3 ATN _T3 ETV _T3 ARID _T3 PFKM _T3 STAT _T3 R3HDM _T3 CDK _T3 CAND _T3 PRDM _T3 N4BP2L _T3 RFC _T3 POSTN _T3 RNASEH2B _T3 DIAPH _T3 PIBF _T3 MYCBP _T3 TPP _T3 TPP _T3 ARHGEF _T3 UPF3A _T3 PRMT _T3 ACIN _T3 ACIN _T3 AP1G _T3 REC _T3 NFKBIA _T3 SOS
118 _T3 DLGAP _T3 DCAF _T3 YLPM _T3 YLPM _T3 GPATCH2L _T3 SPTLC _T3 SMEK _T3 TRIP _T3 CCNK _T3 WARS _T3 DYNC1H _T3 HERC _T3 FAN _T3 EIF2AK _T3 MGA _T3 DMXL _T3 MYO5C _T3 TRIP _T3 UACA _T3 CHD _T3 NDE _T3 XPO _T3 DCTPP _T3 VPS _T3 POLR2C _T3 KATNB _T3 CNOT _T3 GOT _T3 SF3B _T3 WDR _T3 PIEZO
119 _T3 CHMP1A _T3 MYO1C _T3 PRPF _T3 PRPF _T3 POLR2A _T3 CHD _T3 SPECC _T3 MYO18A _T3 LIG _T3 AP2B _T3 CDK _T3 MED _T3 WNK _T3 WNK _T3 DHX _T3 KANSL _T3 LRRC _T3 SPAG _T3 USP _T3 SMURF _T3 TSEN _T3 LLGL _T3 USP _T3 FASN _T3 SLC16A _T3 ROCK _T3 PIK3C _T3 SMAD _T3 VPS4B _T3 PTBP _T3 DAZAP
120 _T3 AP3D _T3 EEF _T3 SAFB _T3 MCOLN _T3 XAB _T3 TIMM _T3 MYO1F _T3 DNM _T3 SMARCA _T3 DOCK _T3 ECSIT _T3 TNPO _T3 UPF _T3 DDX _T3 SIPA1L _T3 FCGBP _T3 ARHGEF _T3 ARHGEF _T3 VASP _T3 DMPK _T3 SYMPK _T3 TRPM _T3 TRPM _T3 TRPM _T3 TMC _T3 ZNF _T3 BIRC _T3 FAM98A _T3 LRPPRC _T3 LRPPRC _T3 EPAS
121 _T3 PSME _T3 TMEM _T3 GCC _T3 RIF _T3 GPD _T3 PKP _T3 NOSTRIN _T3 PPIG _T3 ZAK _T3 HNRNPA _T3 NCKAP _T3 PIKFYVE _T3 ATIC _T3 DIS3L _T3 FARP _T3 NOP _T3 PCED1A _T3 PANK _T3 KIF16B _T3 NXT _T3 CTSA _T3 ZMYND _T3 DDX _T3 PTPN _T3 PCK _T3 DIDO _T3 IFNAR _T3 IL10RB _T3 TTC _T3 MX _T3 MCM3AP
122 _T3 PCNT _T3 HIRA _T3 MAPK _T3 DEPDC _T3 GTPBP _T3 SUN _T3 XRCC _T3 SAMM _T3 TATDN _T3 TOP2B _T3 CLASP _T3 TRANK _T3 LRRFIP _T3 CTNNB _T3 KIF _T3 CDCP _T3 NBEAL _T3 SETD _T3 PLXNB _T3 SLC26A _T3 RAD54L _T3 TEX _T3 PBRM _T3 FLNB _T3 ATXN _T3 UBA _T3 PDIA _T3 SEC61A _T3 RPN _T3 CEP _T3 PLCH
123 _T3 EIF4G _T3 VPS _T3 HTT _T3 RBM _T3 FRYL _T3 OCIAD _T3 SEC31A _T3 LARP _T3 KIAA _T3 OTUD _T3 KLHL _T3 IRF _T3 BRD _T3 TRIP _T3 KIAA _T3 TRIO _T3 ZFR _T3 SKP _T3 HMGCR _T3 RASA _T3 LNPEP _T3 PAM _T3 PPIP5K _T3 HINT _T3 RAPGEF _T3 RAPGEF _T3 CLINT _T3 CDHR _T3 GNB2L _T3 PRRC2A _T3 RING
124 _T3 TAPBP _T3 CUTA _T3 FOXP _T3 ORC _T3 MDN _T3 PTPRK _T3 PTPRK _T3 MED _T3 MTHFD1L _T3 INTS _T3 EIF2AK _T3 HOXA _T3 OGDH _T3 GUSB _T3 DTX _T3 AKAP _T3 ANKIB _T3 SLC25A _T3 BAIAP2L _T3 MCM _T3 GATS _T3 ZNHIT _T3 COG _T3 CHCHD _T3 JHDM1D _T3 BRAF _T3 ZYX _T3 R3HCC _T3 ZNF _T3 MRPL _T3 ASPH _T3 ARFGEF
125 _T3 MAL _T3 ATAD _T3 EPPK _T3 KIAA _T3 NPR _T3 WNK _T3 SLC35D _T3 CIZ _T3 ODF _T3 TOR1A _T3 ANAPC X _T3 MID X _T3 UBA X _T3 HUWE X _T3 FAAH X _T3 DLG X _T3 SLC6A Predicted neoantigens with binding affinity <500nM, allele frequency>5% and mrna expression>10 transcripts per million (TPM).
126 etable 10. Pathways analysis of MHC I and II neoantigens Ingenuity Canonical Group -log(p-value) Ratio Genes MHC Pathways Transcriptional Regulatory FAP 2.76E E-01 SET,TRIM24,RIF1,SKIL,STAT3 MHC_I Network in Embryonic Stem Cells Transcriptional Regulatory Hyper 2.68E E-02 KAT6A MHC_I Network in Embryonic Stem Cells Transcriptional Regulatory Tumor 3.48E E-02 RIF1 MHC_I Network in Embryonic Stem Cells Estrogen Receptor FAP MED13,CDK8,MED20,SPEN,TBP,NCOR1,GTF2F1, MED13L MHC_I Estrogen Receptor MED23 MHC_I Estrogen Receptor Hyper MAPK1,TRRAP,MED14 MHC_I Estrogen Receptor Tumor TAF1,MED23,POLR2A,ERCC3,TRRAP,NCOR1,PC K1,MED24,SMARCA4 MHC_I DNA damage-induced FAP RAD17,CCNB2,ATR MHC_I 3σ DNA damage-induced Tumor AKT2 MHC_I 3σ trna Charging FAP WARS2,EARS2,IARS,FARSB MHC_I trna Charging VARS MHC_I trna Charging Hyper YARS,VARS2,IARS MHC_I trna Charging Tumor WARS,RARS,EPRS MHC_I
127 Protein Ubiquitination Pathway FAP USP21,UBE2A,UBE2Q1,MED20,USP15,HSPH1,UB E3B,USP54,USP30,USP33,USP34,UBE3A MHC_I Protein Ubiquitination Pathway Protein Ubiquitination Pathway PSMD10,USP5,UCHL5,AMFR,SKP2 MHC_I Tumor ANAPC2,USP7,USP15,USP5,PSMD12,BIRC6,ANA MHC_I PC5,UBA1,SKP2 D-myo-inositol-5-phosphate Metabolism FAP PPFIA1,SET,PLCB4,PTPRH,DUSP11,PPP4R1,PPP 3CA,SACM1L MHC_I D-myo-inositol-5-phosphate Metabolism D-myo-inositol-5-phosphate Metabolism D-myo-inositol-5-phosphate Metabolism Cell Cycle: G2/M DNA Damage Checkpoint Regulation Cell Cycle: G2/M DNA Damage Checkpoint Regulation Cell Cycle: G2/M DNA Damage Checkpoint Regulation Cell Cycle: G2/M DNA Damage Checkpoint Regulation PLCD3,PTPRH,DUSP16 MHC_I Hyper MTMR6,SGPP2,PPM1F MHC_I Tumor PTPN1,RASA1,PTEN MHC_I FAP TOP2B,TRIP12,CCNB2,ATR MHC_I Hyper SKP2 MHC_I TRIP12 MHC_I Tumor TOP2B,SKP2 MHC_I ATM FAP HERC2,RAD17,CCNB2,ATR,NBN MHC_I ATM TOPBP1 MHC_I
128 ATM Hyper FANCD2,TRRAP,TLK2,NBN MHC_I ATM Tumor HERC2,USP7,JUN,TOPBP1,TRRAP MHC_I Wnt/β-catenin FAP AXIN2,APPL2,CSNK1A1,TLE1,BMPR2,SOX9,TCF7 L2,LRP1 MHC_I Wnt/β-catenin Hyper TGFBR1,CSNK1G2,MARK2,DVL3,LRP1 MHC_I Wnt/β-catenin Tumor AKT2,JUN,HDAC1,ILK,CTNNB1 MHC_I D-myo-inositol (1,4,5,6)- Tetrakisphosphate Biosynthesis D-myo-inositol (1,4,5,6)- Tetrakisphosphate Biosynthesis D-myo-inositol (1,4,5,6)- Tetrakisphosphate Biosynthesis D-myo-inositol (1,4,5,6)- Tetrakisphosphate Biosynthesis D-myo-inositol (3,4,5,6)- tetrakisphosphate Biosynthesis D-myo-inositol (3,4,5,6)- tetrakisphosphate Biosynthesis D-myo-inositol (3,4,5,6)- tetrakisphosphate Biosynthesis D-myo-inositol (3,4,5,6)- tetrakisphosphate Biosynthesis FAP PPFIA1,SET,PTPRH,DUSP11,PPP4R1,PPP3CA,SA CM1L Hyper MHC_I PTPRH,DUSP16 MHC_I MTMR6,SGPP2,PPM1F MHC_I Tumor PTPN1,RASA1,PTEN MHC_I FAP PPFIA1,SET,PTPRH,DUSP11,PPP4R1,PPP3CA,SA CM1L Hyper MHC_I PTPRH,DUSP16 MHC_I MTMR6,SGPP2,PPM1F MHC_I Tumor PTPN1,RASA1,PTEN MHC_I Isoleucine Degradation I FAP ACAT2,EHHADH MHC_I Telomere Extension by FAP TERF2IP,NBN MHC_I Telomerase
129 Telomere Extension by Hyper NBN MHC_I Telomerase Telomere Extension by Tumor XRCC6 MHC_I Telomerase Glutaryl-CoA Degradation FAP ACAT2,EHHADH MHC_I RhoA FAP ROCK2,ACTR2,BAIAP2,ARHGAP12,MYLK,ARHGA P1 MHC_I RhoA RhoA 3-phosphoinositide Degradation 3-phosphoinositide Degradation 3-phosphoinositide Degradation 3-phosphoinositide Degradation Cell Cycle Control of Chromosomal Replication Cell Cycle Control of Chromosomal Replication Cell Cycle Control of Chromosomal Replication Role of BRCA1 in DNA Damage Response Role of BRCA1 in DNA Damage Response Role of BRCA1 in DNA Damage Response ARHGEF1,PLD1 MHC_I Hyper PTK2,ARHGAP5,BAIAP2,PI4KA,PKN1 MHC_I FAP PPFIA1,SET,PTPRH,DUSP11,PPP4R1,PPP3CA,SA MHC_I CM1L PTPRH,MTM1,DUSP16 MHC_I Hyper MTMR6,SGPP2,PPM1F MHC_I Tumor PTPN1,RASA1,PTEN MHC_I FAP TOP2B,PRIM1,ORC4 MHC_I MCM4 MHC_I Tumor TOP2B,ORC3,POLE,MCM4 MHC_I FAP ABRAXAS1,ATR,SMARCD1,NBN MHC_I Hyper PBRM1,TOPBP1,SMARCD1 MHC_I E2F4,FANCD2,SMARCB1,HLTF,NBN MHC_I
130 Role of BRCA1 in DNA Damage Response Tumor PBRM1,SMARCA2,TOPBP1,MSH6,SMARCA4,MLH 1 MHC_I autophagy FAP ATG5,CTSA,VPS41 MHC_I autophagy ATG9A MHC_I autophagy Hyper VPS33A,CTSF MHC_I autophagy Tumor CTSA,MTOR,PIK3C3 MHC_I Leptin in Obesity FAP PLCB4,PDIA3,PDE3B,STAT3 MHC_I Leptin in Obesity PLCD3,FGFR2 MHC_I Leptin in Obesity Hyper MAPK1,ADCY3,ADCY6,PIK3R2 MHC_I Leptin in Obesity Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I NF-κB Activation by Viruses FAP TBP,ITGA1,EIF2AK2,PRKD3 MHC_I NF-κB Activation by Viruses FGFR2 MHC_I NF-κB Activation by Viruses Hyper MAPK1,PRKCH,PIK3R2 MHC_I NF-κB Activation by Viruses Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I Protein Kinase A FAP ROCK2,PLCB4,PTPRH,ADD3,PDIA3,PDE3B,DUSP 11,MYLK,PYGL,PRKD3,PPP3CA,TCF7L2 MHC_I Protein Kinase A PLCD3,PTPRH,PTPRR,DUSP16 MHC_I
131 Protein Kinase A Hyper PTK2,TGFBR1,MAPK1,ITPR2,ADCY3,ADCY6,PRK CH,PTPN3,NFATC4,PTPN21 MHC_I Protein Kinase A Tumor BRAF,ANAPC2,FLNB,PHKB,PTPRK,PTPN1,ITPR3, ANAPC5,PYGB,CTNNB1,PTEN MHC_I Gap Junction FAP PLCB4,PDIA3,TJP1,CSNK1A1,PRKD3,PPP3CA MHC_I Gap Junction Gap Junction Hyper PLCD3,FGFR2 MHC_I MAPK1,CSNK1G2,ITPR2,ADCY3,ADCY6,PRKCH,P IK3R2 MHC_I Gap Junction Tumor AKT2,PIK3C2A,TUBB4B,PIK3C3,PIK3R1,ITPR3,CT NNB1,NPR2 MHC_I Pyridoxal 5'-phosphate Salvage Pathway Pyridoxal 5'-phosphate Salvage Pathway Pyridoxal 5'-phosphate Salvage Pathway FAP CDK8,CSNK1A1,EIF2AK2 MHC_I Hyper PRKX,DMPK,MAPK1,PRKCH,PKN1 MHC_I Tumor BRAF,AKT2,DMPK MHC_I TR/RXR Activation FAP PDE3B,NCOA6,NCOR1,THRA MHC_I TR/RXR Activation FGFR2 MHC_I TR/RXR Activation Tumor SLC16A3,AKT2,MTOR,PIK3C2A,PIK3C3,PIK3R1,F ASN,NCOR1,PCK1 MHC_I
132 Mouse Embryonic Stem Cell Pluripotency Mouse Embryonic Stem Cell Pluripotency Mouse Embryonic Stem Cell Pluripotency Mouse Embryonic Stem Cell Pluripotency Epithelial Adherens Junction Epithelial Adherens Junction FAP IL6ST,BMPR2,STAT3,TCF7L2 MHC_I FGFR2 MHC_I Hyper MAPK1,DVL3,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,CTNNB1 MHC_I FAP ACTR2,BAIAP2,BMPR2,TCF7L2,CLIP1 MHC_I Hyper RAPGEF1,MYH9,TGFBR1,NECTIN3,BAIAP2,ACTN 1,NECTIN2 MHC_I Epithelial Adherens Junction Tumor AKT2,TUBB4B,NECTIN3,ZYX,CLINT1,CTNNB1,CT NND1,PTEN MHC_I Stearate Biosynthesis I (Animals) Stearate Biosynthesis I (Animals) FAP ZADH2,ELOVL1 MHC_I LPCAT4 MHC_I Hyper SRD5A3,THEM4 MHC_I Tumor ACSL3,FASN,ACOT9 MHC_I Stearate Biosynthesis I (Animals) Stearate Biosynthesis I (Animals) RAR Activation FAP TRIM24,RDH14,NCOR1,SMARCD1,RDH13,PRKD3 MHC_I RAR Activation RAR Activation Hyper PBRM1,STAT5A,SMARCD1 MHC_I STAT5A,MAPK1,SMARCB1,ADCY3,ADCY6,PRKCH,PIK3R2,HLTF,PSMC5 MHC_I RAR Activation Tumor PBRM1,AKT2,JUN,SMARCA2,ERCC3,PIK3R1,NCO MHC_I
133 R1,SMARCA4,PTEN Cleavage and Polyadenylation of Pre-mRNA Cleavage and Polyadenylation of Pre-mRNA Cleavage and Polyadenylation of Pre-mRNA Role of NFAT in Cardiac Hypertrophy FAP CSTF1 MHC_I CPSF2 MHC_I Hyper CPSF1 MHC_I FAP IL6ST,PLCB4,PDIA3,CSNK1A1,PRKD3,PPP3CA MHC_I Role of NFAT in Cardiac Hypertrophy Role of NFAT in Cardiac Hypertrophy Hyper PLCD3,FGFR2 MHC_I TGFBR1,CAMK1,MAPK1,HDAC8,ITPR2,ADCY3,AD CY6,PRKCH,PIK3R2,NFATC4 MHC_I Role of NFAT in Cardiac Hypertrophy Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,HDAC1,ITPR3,CAB IN1 MHC_I Cholesterol Biosynthesis I FAP DHCR7 MHC_I Cholesterol Biosynthesis I Tumor HSD17B7,LBR MHC_I Cholesterol Biosynthesis II FAP DHCR7 MHC_I (via 24,25-dihydrolanosterol) Cholesterol Biosynthesis II Tumor HSD17B7,LBR MHC_I (via 24,25-dihydrolanosterol) Cholesterol Biosynthesis III FAP DHCR7 MHC_I (via Desmosterol) Cholesterol Biosynthesis III Tumor HSD17B7,LBR MHC_I (via Desmosterol) Gαq FAP ROCK2,BTK,PLCB4,PRKD3,PPP3CA MHC_I Gαq FGFR2,PLD1 MHC_I
134 Gαq Hyper MAPK1,ITPR2,PRKCH,PIK3R2,NFATC4 MHC_I Gαq Tumor AKT2,PLD3,PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency FAP IL6ST,RIF1,BMPR2,STAT3 MHC_I FGFR2 MHC_I Tumor AKT2,RIF1,PIK3C2A,PIK3C3,PIK3R1,CTNNB1 MHC_I IL-3 FAP STAT3,PRKD3,PPP3CA MHC_I IL STAT5A,FGFR2 MHC_I IL-3 Hyper RAPGEF1,STAT5A,MAPK1,PRKCH,PIK3R2 MHC_I IL-3 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Thrombin FAP ROCK2,PLCB4,PDIA3,TBP,MYLK,PRKD3 MHC_I Thrombin Thrombin Hyper PLCD3,FGFR2,ARHGEF1 MHC_I PTK2,CAMK1,MAPK1,ITPR2,ADCY3,ADCY6,PRKC H,PIK3R2 MHC_I Thrombin Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,ITPR3,ARHGEF1 MHC_I PI3K in B Lymphocytes PI3K in B Lymphocytes FAP BTK,PLCB4,PDIA3,PPP3CA MHC_I PLCD3 MHC_I
135 PI3K in B Lymphocytes PI3K in B Lymphocytes Assembly of RNA Polymerase II Complex Assembly of RNA Polymerase II Complex Hyper MAPK1,ITPR2,PIK3R2,NFATC4 MHC_I Tumor AKT2,JUN,PIK3R1,ITPR3,PTEN MHC_I FAP TBP,GTF2F1 MHC_I Tumor TAF1,POLR2A,ERCC3 MHC_I PPARα/RXRα Activation FAP PLCB4,PDIA3,NCOA6,BMPR2,NCOR1 MHC_I PPARα/RXRα Activation PLCD3,MED23,CPT1B MHC_I PPARα/RXRα Activation Hyper TGFBR1,MAPK1,ADCY3,ADCY6 MHC_I PPARα/RXRα Activation Tumor CAND1,MED23,JUN,GPD2,FASN,NCOR1,MED24,A DIPOR1 MHC_I Acute Myeloid Leukemia Acute Myeloid Leukemia Acute Myeloid Leukemia Acute Myeloid Leukemia FAP STAT3,SPI1,TCF7L2 MHC_I STAT5A,FGFR2 MHC_I Hyper STAT5A,MAPK1,PIK3R2 MHC_I Tumor BRAF,AKT2,MTOR,PIK3C2A,PIK3C3,PIK3R1 MHC_I p70s6k FAP BTK,PLCB4,PDIA3,PRKD3 MHC_I p70s6k PLCD3,FGFR2,PLD1 MHC_I p70s6k Hyper MAPK1,PRKCH,PIK3R2,BCAP31 MHC_I p70s6k Tumor AKT2,MTOR,PIK3C2A,EEF2,PIK3C3,PIK3R1 MHC_I
136 EIF2 FAP PTBP1,EIF5B,EIF4G3,EIF2AK2,RPS17,EIF2AK3 MHC_I EIF2 Tumor EIF2AK4,AKT2,WARS,PIK3C2A,PIK3C3,PIK3R1,EI F5B MHC_I Phagosome Maturation FAP DYNC1H1,CTSA,VPS41,ATP6V0D1 MHC_I Phagosome Maturation ATP6V0A2,ATP6V0D1 MHC_I Phagosome Maturation Hyper CALR,VPS33A,NAPA,CTSF MHC_I Phagosome Maturation Tumor DYNC1H1,CTSA,TUBB4B,PIK3C3,ATP6V0D1,ATP6 V0A1 MHC_I Salvage Pathways of Pyrimidine Ribonucleotides Salvage Pathways of Pyrimidine Ribonucleotides Salvage Pathways of Pyrimidine Ribonucleotides DNA Methylation and Transcriptional Repression DNA Methylation and Transcriptional Repression FAP CDK8,CSNK1A1,EIF2AK2 MHC_I Hyper PRKX,DMPK,MAPK1,PRKCH,UCKL1,PKN1 MHC_I Tumor BRAF,AKT2,DMPK,PCK1 MHC_I FAP DNMT1 MHC_I Tumor CHD3,HDAC1 MHC_I CNTF FAP IL6ST,STAT3 MHC_I CNTF FGFR2 MHC_I CNTF Hyper MAPK1,PIK3R2 MHC_I CNTF Tumor MTOR,PIK3C2A,PIK3C3,PIK3R1 MHC_I UVA-Induced MAPK FAP PLCB4,PDIA3,PARP4 MHC_I
137 UVA-Induced MAPK PLCD3,FGFR2,SMPD1,PARP9 MHC_I UVA-Induced MAPK Hyper MAPK1,PIK3R2 MHC_I UVA-Induced MAPK Tumor MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1,PARP14 MHC_I Lipid Antigen Presentation by FAP PDIA3 MHC_I CD1 Lipid Antigen Presentation by Hyper CALR MHC_I CD1 Lipid Antigen Presentation by Tumor AP1G2,AP2B1 MHC_I CD1 Role of JAK1, JAK2 and TYK2 FAP STAT3 MHC_I in Interferon Role of JAK1, JAK2 and TYK2 Tumor IFNGR2,IFNAR2 MHC_I in Interferon Thrombopoietin FAP STAT3,PRKD3 MHC_I Thrombopoietin STAT5A,FGFR2 MHC_I Thrombopoietin Hyper STAT5A,MAPK1,PRKCH,PIK3R2 MHC_I Thrombopoietin Tumor JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I T Cell Receptor FAP BTK,PTPRH,PPP3CA MHC_I T Cell Receptor PTPRH,FGFR2 MHC_I T Cell Receptor Hyper MAPK1,PIK3R2,NFATC4 MHC_I T Cell Receptor Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,RASA1 MHC_I Remodeling of Epithelial Adherens Junctions Remodeling of Epithelial Adherens Junctions FAP ACTR2,CLIP1 MHC_I Hyper DNM1,HGS,ACTN1 MHC_I
138 Remodeling of Epithelial Adherens Junctions Neuropathic Pain In Dorsal Horn Neurons Neuropathic Pain In Dorsal Horn Neurons Neuropathic Pain In Dorsal Horn Neurons Neuropathic Pain In Dorsal Horn Neurons Tumor TUBB4B,ZYX,CTNNB1,CTNND1 MHC_I FAP PLCB4,PDIA3,PRKD3 MHC_I PLCD3,FGFR2 MHC_I Hyper CAMK1,MAPK1,ITPR2,PRKCH,PIK3R2 MHC_I Tumor PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I HGF FAP ELF3,STAT3,PRKD3 MHC_I HGF ELF4,FGFR2 MHC_I HGF Hyper PTK2,RAPGEF1,MAP3K11,MAPK1,PRKCH,PIK3R2 MHC_I HGF Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I GM-CSF FAP STAT3,PPP3CA MHC_I GM-CSF FGFR2 MHC_I GM-CSF Hyper MAPK1,PIK3R2 MHC_I GM-CSF Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I Tight Junction FAP EPB41,CSTF1,TJP1,MYLK MHC_I Tight Junction Hyper EPB41,MYH9,TGFBR1,TNFRSF1A,NECTIN3,MARK 2,CPSF1,CASK,NAPA,NECTIN2 MHC_I Tight Junction Tumor AKT2,JUN,NECTIN3,STX4,CTNNB1,PTEN MHC_I mediated FAP PLCB4,PDIA3,PRKD3 MHC_I
139 mediated mediated Hyper PLCD3,FGFR2 MHC_I TSC1,MAPK1,TNFRSF1A,TSC2,PRKCH,PIK3R2 MHC_I mediated Tumor AKT2,JUN,PIK3C2A,TUBB4B,PIK3C3,PIK3R1 MHC_I Prolactin FAP STAT3,PRKD3 MHC_I Prolactin STAT5A,FGFR2 MHC_I Prolactin Hyper STAT5A,MAPK1,PRKCH,PIK3R2 MHC_I Prolactin Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,IRF1 MHC_I JAK/Stat FAP PIAS2,STAT3 MHC_I JAK/Stat STAT5A,FGFR2 MHC_I JAK/Stat Hyper STAT5A,MAPK1,PIK3R2 MHC_I JAK/Stat Tumor AKT2,MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1,PTPN1 MHC_I P2Y Purigenic Receptor Pathway P2Y Purigenic Receptor Pathway P2Y Purigenic Receptor Pathway P2Y Purigenic Receptor Pathway Role of NFAT in Regulation of the Immune Response FAP PLCB4,PDIA3,PRKD3 MHC_I PLCD3,FGFR2 MHC_I Hyper MAPK1,ADCY3,ADCY6,PRKCH,PIK3R2 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I FAP BTK,PLCB4,CSNK1A1,PPP3CA MHC_I
140 Role of NFAT in Regulation of the Immune Response Role of NFAT in Regulation of the Immune Response Hyper MAPK1,CSNK1G2,ITPR2,PIK3R2,NFATC4 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3,CABIN1 MHC_I PDGF FAP STAT3,EIF2AK2 MHC_I PDGF FGFR2 MHC_I PDGF Hyper MAPK1,PIK3R2 MHC_I PDGF Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,RASA1 MHC_I IL-9 FAP STAT3 MHC_I IL STAT5A,FGFR2 MHC_I IL-9 Hyper STAT5A,PIK3R2 MHC_I IL-9 Tumor PIK3C2A,PIK3C3,PIK3R1 MHC_I Glucocorticoid Receptor Glucocorticoid Receptor Hyper PBRM1,STAT5A,FGFR2,SMARCD1 MHC_I STAT5A,TGFBR1,MAPK1,SMARCB1,PIK3R2,NFAT C4,HLTF,MED14 MHC_I Glucocorticoid Receptor Tumor PBRM1,AKT2,PIK3C2A,PIK3R1,PCK1,SMARCA4,J UN,POLR2A,TAF1,SMARCA2,ERCC3,PIK3C3,NCO R1 MHC_I Neuregulin Neuregulin Hyper STAT5A MHC_I STAT5A,MAPK1,PRKCH,ERBB2,PIK3R2 MHC_I
141 Neuregulin Tumor AKT2,MTOR,PIK3R1,PTEN MHC_I Actin Cytoskeleton Actin Cytoskeleton Hyper DIAPH2,FGFR2,ARHGEF1 MHC_I PTK2,MYH9,WASF2,MAPK1,BAIAP2,PIK3R2,GIT1, ACTN1 MHC_I Actin Cytoskeleton Tumor PIK3C2A,PIK3C3,PIK3R1,TRIO,ARHGEF1,NCKAP1 MHC_I Huntington's Disease Hyper DNM1,MAPK1,HDAC8,CASP2,PSME4,PRKCH,PIK3 R2,NAPA MHC_I Huntington's Disease Tumor AKT2,PIK3C2A,PIK3R1,HDAC1,APAF1,MTOR,JUN, POLR2A,PIK3C3,HTT,PSME4,TCERG1,NCOR1,RA SA1,CASP5 MHC_I p TOPBP1,FGFR2 MHC_I p53 Hyper CCNG1,SIRT1,PIK3R2 MHC_I p53 Tumor AKT2,JUN,PIK3C2A,TOPBP1,PIK3C3,PIK3R1,HDA C1,APAF1,CTNNB1,PTEN MHC_I Erythropoietin STAT5A,FGFR2 MHC_I Erythropoietin Hyper STAT5A,MAPK1,PRKCH,PIK3R2 MHC_I Erythropoietin Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Fcγ Receptor-mediated Phagocytosis in Macrophages PLD1 MHC_I
142 and Monocytes Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes IL-8 IL-8 Hyper MAPK1,PRKCH,PIK3R2 MHC_I Tumor AKT2,PLD3,PIK3R1,PTEN MHC_I Hyper FGFR2,PLD1 MHC_I PTK2,MAPK1,PRKCH,PIK3R2 MHC_I IL-8 Tumor BRAF,AKT2,ARRB2,MTOR,PLD3,JUN,PIK3C2A,PIK 3C3,PIK3R1 MHC_I IL-12 and Production in Macrophages IL-12 and Production in Macrophages IL-12 and Production in Macrophages Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses FcγRIIB in B Lymphocytes FcγRIIB in B Lymphocytes LPS-stimulated MAPK FGFR2,MST1R MHC_I Hyper MAPK1,PRKCH,PIK3R2 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,IRF1 MHC_I Hyper OAS2,FGFR2,TLR3 MHC_I MAPK1,PRKCH,PIK3R2,OAS3 MHC_I Tumor PIK3C2A,PIK3C3,PIK3R1,C1QA MHC_I FGFR2 MHC_I Tumor PIK3C2A,PIK3C3,PIK3R1 MHC_I FGFR2 MHC_I
143 LPS-stimulated MAPK LPS-stimulated MAPK CD40 CD40 Hyper MAPK1,PRKCH,PIK3R2 MHC_I Tumor JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Hyper FGFR2 MHC_I MAPK1,PIK3R2 MHC_I CD40 Tumor JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I CXCR4 Hyper PTK2,MAPK1,ITPR2,ADCY3,ADCY6,PRKCH,PIK3R 2 MHC_I CXCR4 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I CD28 in T Helper Cells CD28 in T Helper Cells CD28 in T Helper Cells IL-15 IL FGFR2 MHC_I Hyper ITPR2,PIK3R2,NFATC4 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I Hyper STAT5A,FGFR2 MHC_I PTK2,STAT5A,MAPK1,PIK3R2 MHC_I IL-15 Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I Virus Entry via Endocytic Pathways Virus Entry via Endocytic Pathways Hyper FGFR2 MHC_I DNM1,PRKCH,PIK3R2 MHC_I
144 Virus Entry via Endocytic Pathways Reelin in Neurons Reelin in Neurons Tumor FLNB,AP1G2,PIK3C2A,PIK3C3,PIK3R1,AP2B1 MHC_I Hyper FGFR2,ARHGEF1 MHC_I MAP3K11,PIK3R2 MHC_I Reelin in Neurons Tumor PIK3C2A,PIK3C3,PIK3R1,ARHGEF1 MHC_I Endothelin-1 Endothelin-1 Hyper PLCD3,FGFR2,PLD1 MHC_I MAPK1,ITPR2,CASP2,ADCY3,ADCY6,PRKCH,PIK3 R2 MHC_I Endothelin-1 Tumor BRAF,PLD3,JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3,C ASP5 MHC_I Relaxin FGFR2 MHC_I Relaxin Hyper MAPK1,ADCY3,ADCY6,PIK3R2 MHC_I Relaxin Tumor BRAF,AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,NPR2 MHC_I Renin-Angiotensin Renin-Angiotensin Hyper FGFR2 MHC_I PTK2,MAPK1,ITPR2,ADCY3,ADCY6,PRKCH,PIK3R 2 MHC_I Renin-Angiotensin Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I icos-icosl in T Helper Cells FGFR2 MHC_I
145 icos-icosl in T Helper Cells icos-icosl in T Helper Cells Molecular Mechanisms of Cancer Molecular Mechanisms of Cancer Hyper ITPR2,PIK3R2,NFATC4 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,ITPR3,PTEN MHC_I Hyper ZBTB17,FGFR2,ARHGEF1 MHC_I PTK2,RAPGEF1,E2F4,TGFBR1,MAPK1,FANCD2,A DCY3,ADCY6,PRKCH,PIK3R2,LRP1,NBN MHC_I Molecular Mechanisms of Cancer Tumor BRAF,AKT2,JUN,TFDP1,PIK3C2A,PIK3C3,PIK3R1, APAF1,ARHGEF1,CTNNB1,RASA1,CTNND1 MHC_I FLT3 in Hematopoietic Progenitor Cells FLT3 in Hematopoietic Progenitor Cells FLT3 in Hematopoietic Progenitor Cells GNRH Hyper STAT5A,FGFR2 MHC_I STAT5A,MAPK1,PIK3R2 MHC_I Tumor AKT2,MTOR,PIK3C2A,PIK3C3,PIK3R1 MHC_I Hyper PTK2,DNM1,MAP3K11,MAPK1,ITPR2,ADCY3,ADC Y6,PRKCH MHC_I GNRH Tumor JUN,ITPR3 MHC_I Germ Cell-Sertoli Cell Hyper PTK2,TGFBR1,MAP3K11,MAPK1,TNFRSF1A,NECT MHC_I Junction IN3,PIK3R2,ACTN1,NECTIN2
146 Germ Cell-Sertoli Cell Junction Tumor PIK3C2A,TUBB4B,PIK3C3,PIK3R1,NECTIN3,ILK,ZY X,CLINT1,CTNNB1,CTNND1 MHC_I Glioma FGFR2 MHC_I Glioma Hyper E2F4,CAMK1,MAPK1,PRKCH,PIK3R2 MHC_I Glioma Tumor AKT2,MTOR,TFDP1,PIK3C2A,PIK3C3,PIK3R1,PTE N MHC_I Type II Diabetes Mellitus Type II Diabetes Mellitus Type II Diabetes Mellitus FGFR2,SMPD1 MHC_I Hyper MAPK1,TNFRSF1A,PRKCH,PIK3R2 MHC_I Tumor AKT2,ACSL3,MTOR,PIK3C2A,PIK3C3,PIK3R1,ADIP MHC_I OR1 Non-Small Cell Lung Cancer Non-Small Cell Lung Cancer Non-Small Cell Lung Cancer Production of Nitric Oxide and Reactive Oxygen Species in Macrophages Production of Nitric Oxide and Reactive Oxygen Species in Macrophages FGFR2 MHC_I Hyper MAPK1,ITPR2,ERBB2,PIK3R2 MHC_I Tumor AKT2,TFDP1,PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I Hyper MAP3K11,MAPK1,TNFRSF1A,PRKCH,PIK3R2 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,IFNGR2,IRF1 MHC_I Gα12/ FGFR2,ARHGEF1 MHC_I Gα12/13 Hyper PTK2,MAPK1,PIK3R2 MHC_I
147 Gα12/13 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,ARHGEF1,CT NNB1,RASA1 MHC_I Colorectal Cancer Metastasis Colorectal Cancer Metastasis Hyper FGFR2,TLR3 MHC_I E2F4,TGFBR1,MAPK1,TNFRSF1A,ADCY3,ADCY6, PIK3R2,LRP1 MHC_I Colorectal Cancer Metastasis Tumor BRAF,AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,MSH6,C TNNB1,MLH1 MHC_I Pancreatic Adenocarcinoma Pancreatic Adenocarcinoma Pancreatic Adenocarcinoma Sphingosine-1-phosphate Sphingosine-1-phosphate Sphingosine-1-phosphate Systemic Lupus Erythematosus Systemic Lupus Erythematosus FGFR2,PLD1 MHC_I Hyper E2F4,TGFBR1,MAPK1,ERBB2,PIK3R2 MHC_I Tumor AKT2,PLD3,TFDP1,PIK3C2A,PIK3C3,PIK3R1 MHC_I PLCD3,FGFR2,SMPD1 MHC_I Hyper PTK2,MAPK1,CASP2,ADCY3,ADCY6,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,CASP5 MHC_I EFTUD2,FGFR2 MHC_I Tumor AKT2,MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1,PRPF8, MHC_I SNRNP200 AMPK PBRM1,CPT1A,CPT1B,FGFR2,SMARCD1 MHC_I
148 AMPK Hyper TSC1,MAPK1,SMARCB1,SIRT1,TSC2,PIK3R2,M T8,HLTF MHC_I AMPK Tumor PBRM1,AKT2,MTOR,PIK3C2A,EEF2,SMARCA2,PIK 3C3,PIK3R1,FASN,SMARCA4,PFKM MHC_I PAK FGFR2 MHC_I PAK Hyper PTK2,MAPK1,EPHB3,PIK3R2,GIT1 MHC_I PAK Tumor PIK3C2A,PIK3C3,PIK3R1 MHC_I Hereditary Breast Cancer Hereditary Breast Cancer Hereditary Breast Cancer PBRM1,FGFR2,SMARCD1 MHC_I Hyper HDAC8,FANCD2,SMARCB1,PIK3R2,HLTF,NBN MHC_I Tumor PBRM1,AKT2,POLR2A,PIK3C2A,SMARCA2,PIK3C MHC_I 3,PIK3R1,HDAC1,MSH6,SMARCA4,MLH1,PTEN Rac Rac Hyper FGFR2,PLD1 MHC_I PTK2,MAP3K11,MAPK1,BAIAP2,PIK3R2,PI4KA MHC_I Rac Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,NCKAP1 MHC_I Ovarian Cancer FGFR2 MHC_I
149 Ovarian Cancer Tumor BRAF,AKT2,MTOR,TFDP1,PIK3C2A,PIK3C3,PIK3R 1,MSH6,CTNNB1,MLH1,PTEN MHC_I HER-2 in Breast Cancer HER-2 in Breast Cancer HER-2 in Breast Cancer Breast Cancer Regulation by Stathmin1 Breast Cancer Regulation by Stathmin FGFR2 MHC_I Hyper TSC1,TSC2,PRKCH,ERBB2,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I Hyper FGFR2,ARHGEF1 MHC_I E2F4,CAMK1,MAPK1,ITPR2,ADCY3,ADCY6,PRKC H,PIK3R2 MHC_I Breast Cancer Regulation by Stathmin1 Tumor PIK3C2A,TUBB4B,PIK3C3,PIK3R1,ITPR3,ARHGEF 1 MHC_I RANK in Osteoclasts RANK in Osteoclasts RANK in Osteoclasts Glioblastoma Multiforme Glioblastoma Multiforme Glioblastoma Multiforme Nur77 in T Lymphocytes FGFR2 MHC_I Hyper MAP3K11,MAPK1,PIK3R2 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I PLCD3,FGFR2 MHC_I Hyper TSC1,E2F4,MAPK1,ITPR2,TSC2,PIK3R2 MHC_I Tumor AKT2,MTOR,PIK3C2A,PIK3C3,PIK3R1,ITPR3,CTN MHC_I NB1,PTEN Tumor HDAC1,APAF1,CABIN1 MHC_I
150 IL-17A in Airway Cells IL-17A in Airway Cells IL-17A in Airway Cells NGF NGF FGFR2 MHC_I Hyper MAPK1,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,MUC5B,PTEN MHC_I Hyper FGFR2,SMPD1 MHC_I MAP3K11,MAPK1,PIK3R2 MHC_I NGF Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,TRIO MHC_I Sertoli Cell-Sertoli Cell Junction Hyper EPB41,MAP3K11,MAPK1,TNFRSF1A,NECTIN3,AC TN1,NECTIN2 MHC_I Sertoli Cell-Sertoli Cell Junction Tumor AKT2,JUN,TUBB4B,NECTIN3,ILK,CLINT1,CTNNB1, PTEN MHC_I Telomerase ELF4,FGFR2 MHC_I Telomerase Hyper MAPK1,HDAC8,TEP1,PIK3R2 MHC_I Telomerase Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,HDAC1 MHC_I enos FGFR2 MHC_I enos Hyper NOSIP,ITPR2,ADCY3,ADCY6,PRKCH,PIK3R2 MHC_I enos Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,ITPR3,NOSTRIN MHC_I VEGF Family Ligand- Receptor Interactions FGFR2 MHC_I
151 VEGF Family Ligand- Receptor Interactions VEGF Family Ligand- Receptor Interactions Ephrin A Ephrin A Hyper MAPK1,PRKCH,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I Hyper FGFR2 MHC_I PTK2,PIK3R2,EPHA2 MHC_I Ephrin A Tumor PIK3C2A,PIK3C3,PIK3R1,EPHA2 MHC_I ErbB FGFR2 MHC_I ErbB Hyper MAPK1,PRKCH,ERBB2,PIK3R2 MHC_I ErbB Tumor MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I ErbB2-ErbB STAT5A,FGFR2 MHC_I ErbB2-ErbB3 Hyper STAT5A,MAPK1,ERBB2,PIK3R2 MHC_I ErbB2-ErbB3 Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,PTEN MHC_I PEDF FGFR2 MHC_I PEDF Hyper HNF1B,SOD2,WASF2,MAPK1,PIK3R2,TCF12 MHC_I PEDF Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I UVB-Induced MAPK FGFR2 MHC_I
152 UVB-Induced MAPK Hyper MAPK1,PRKCH,PIK3R2 MHC_I UVB-Induced MAPK Tumor MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I EGF FGFR2 MHC_I EGF Hyper MAPK1,ITPR2,PIK3R2 MHC_I EGF Tumor AKT2,MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3, RASA1 MHC_I SAPK/JNK FGFR2 MHC_I SAPK/JNK Hyper MINK1,MAP3K11,PIK3R2 MHC_I SAPK/JNK Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,MAP4K5 MHC_I Nitric Oxide in the Cardiovascular System Nitric Oxide in the Cardiovascular System Nitric Oxide in the Cardiovascular System FGF FGF FGFR2 MHC_I Hyper MAPK1,ITPR2,ATP2A3,PRKCH,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,ITPR3 MHC_I Hyper FGFR2 MHC_I MAPK1,PIK3R2 MHC_I FGF Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1 MHC_I B Cell Receptor Hyper PTK2,MAP3K11,MAPK1,PIK3R2,NFATC4 MHC_I
153 B Cell Receptor Tumor AKT2,MTOR,JUN,PIK3C2A,PIK3C3,PIK3R1,PTEN MHC_I Insulin Receptor FGFR2 MHC_I Insulin Receptor Hyper TSC1,RAPGEF1,MAPK1,TSC2,PIK3R2 MHC_I Insulin Receptor Tumor AKT2,MTOR,PIK3C2A,PIK3C3,PIK3R1,PTPN1,STX 4,PTEN MHC_I Integrin Hyper PTK2,ARHGAP5,RAPGEF1,MAP3K11,MAPK1,PIK3 R2,GIT1,ACTN1 MHC_I Integrin Tumor BRAF,PARVB,AKT2,PIK3C2A,PIK3C3,PIK3R1,ILK,Z YX,PTEN MHC_I IGF FGFR2 MHC_I IGF-1 Hyper PTK2,MAPK1,PIK3R2 MHC_I IGF-1 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,RASA1 MHC_I NF-κB FGFR2,TLR3 MHC_I NF-κB Tumor BRAF,AKT2,PIK3C2A,PIK3C3,PIK3R1,HDAC1 MHC_I IL FGFR2 MHC_I IL-6 Hyper MAPK1,TNFRSF1A,PIK3R2 MHC_I IL-6 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Th1 Pathway FGFR2 MHC_I
154 Th1 Pathway Tumor PIK3C2A,PIK3C3,PIK3R1,IFNGR2,IRF1 MHC_I Calcium Hyper CALR,MYH9,CAMK1,MAPK1,HDAC8,ITPR2,ATP2A 3,ASPH,NFATC4 MHC_I Calcium Tumor HDAC1,ITPR3,ASPH,CABIN1 MHC_I Ceramide FGFR2,SMPD1 MHC_I Ceramide Hyper TNFRSF1A,PIK3R2 MHC_I Ceramide Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Lymphotoxin β Receptor Lymphotoxin β Receptor Lymphotoxin β Receptor IL-17 IL FGFR2 MHC_I Hyper MAPK1,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,APAF1 MHC_I Hyper FGFR2 MHC_I MAPK1,PIK3R2 MHC_I IL-17 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,MUC5B MHC_I CTLA4 in Cytotoxic T Lymphocytes CTLA4 in Cytotoxic T Lymphocytes IL-15 Production FGFR2 MHC_I Tumor AKT2,AP1G2,PIK3C2A,PIK3C3,PIK3R1,AP2B1 MHC_I MST1R MHC_I
155 IL-15 Production Hyper PTK2,MAP3K11 MHC_I IL-15 Production Tumor IRF1 MHC_I HIF1α EGLN3,FGFR2 MHC_I HIF1α Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Angiopoietin STAT5A,FGFR2 MHC_I Angiopoietin Hyper PTK2,STAT5A,PIK3R2 MHC_I Angiopoietin Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,RASA1 MHC_I Docosahexaenoic Acid (DHA) Docosahexaenoic Acid (DHA) Docosahexaenoic Acid (DHA) HMGB1 HMGB FGFR2 MHC_I Hyper PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,APAF1 MHC_I Hyper FGFR2 MHC_I MAPK1,TNFRSF1A,KAT6A,PIK3R2,KAT2A MHC_I HMGB1 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,IFNGR2 MHC_I Polyamine Regulation in Colon Cancer Polyamine Regulation in Colon Cancer Antiproliferative Role of Somatostatin Receptor 2 Antiproliferative Role of Somatostatin Receptor 2 Hyper PSME4 MHC_I Tumor PSME4,CTNNB1 MHC_I Hyper FGFR2 MHC_I MAPK1,PIK3R2 MHC_I
156 Antiproliferative Role of Somatostatin Receptor 2 Melanoma Melanoma Tumor BRAF,PIK3C2A,PIK3C3,PIK3R1,NPR2 MHC_I Hyper FGFR2 MHC_I MAPK1,PIK3R2 MHC_I Melanoma Tumor BRAF,AKT2,PIK3C2A,PIK3C3,PIK3R1,PTEN MHC_I Prostate Cancer FGFR2 MHC_I Prostate Cancer Hyper MAPK1,PIK3R2 MHC_I Prostate Cancer Tumor AKT2,MTOR,TFDP1,PIK3C2A,PIK3C3,PIK3R1,CTN NB1,PTEN MHC_I Renal Cell Carcinoma Renal Cell Carcinoma Renal Cell Carcinoma Small Cell Lung Cancer Small Cell Lung Cancer Small Cell Lung Cancer EGLN3,FGFR2 MHC_I Hyper RAPGEF1,MAPK1,PIK3R2 MHC_I Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I FGFR2,SKP2 MHC_I Hyper PTK2,PIK3R2 MHC_I Tumor AKT2,TFDP1,PIK3C2A,PIK3C3,PIK3R1,APAF1,SKP MHC_I 2,PTEN Endometrial Cancer Endometrial Cancer Hyper FGFR2 MHC_I MAPK1,ERBB2,PIK3R2 MHC_I
157 Endometrial Cancer Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,ILK,CTNNB1,MLH1, PTEN MHC_I Chronic Myeloid Leukemia Chronic Myeloid Leukemia Hyper STAT5A,FGFR2 MHC_I STAT5A,E2F4,TGFBR1,MAPK1,HDAC8,PIK3R2 MHC_I Chronic Myeloid Leukemia Myc Mediated Apoptosis Myc Mediated Apoptosis ILK Tumor AKT2,TFDP1,PIK3C2A,PIK3C3,PIK3R1,HDAC1 MHC_I FGFR2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,APAF1 MHC_I Hyper PTK2,MYH9,MAPK1,TNFRSF1A,PIK3R2,ACTN1 MHC_I ILK Tumor PARVB,FLNB,AKT2,MTOR,JUN,PIK3C2A,PIK3C3,P IK3R1,ILK,CTNNB1,PTEN MHC_I FAK FGFR2 MHC_I FAK Hyper PTK2,MAPK1,PIK3R2 MHC_I FAK Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,PTEN MHC_I Estrogen-Dependent Breast Cancer Estrogen-Dependent Breast Cancer Hyper STAT5A,FGFR2 MHC_I STAT5A,MAPK1,PIK3R2 MHC_I
158 Estrogen-Dependent Breast Cancer Role of PI3K/AKT in the Pathogenesis of Influenza Role of PI3K/AKT in the Pathogenesis of Influenza Role of PI3K/AKT in the Pathogenesis of Influenza Antiproliferative Role of TOB in T Cell Antiproliferative Role of TOB in T Cell Antiproliferative Role of TOB in T Cell Inhibition of Angiogenesis by TSP1 Inhibition of Angiogenesis by TSP1 Mismatch Repair in Eukaryotes Role of Tissue Factor in Cancer Role of Tissue Factor in Cancer Role of Tissue Factor in Cancer Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1,HSD17B7 MHC_I FGFR2 MHC_I Hyper MAPK1,PIK3R2 MHC_I Tumor AKT2,PIK3C2A,PIK3C3,PIK3R1,MLH1 MHC_I SKP2 MHC_I Hyper TGFBR1,MAPK1 MHC_I Tumor SKP2 MHC_I Hyper HSPG2,TGFBR1,MAPK1 MHC_I Tumor AKT2,JUN MHC_I Tumor MSH6,MLH1 MHC_I STAT5A,FGFR2 MHC_I Hyper STAT5A,MAPK1,PIK3R2 MHC_I Tumor AKT2,ARRB2,MTOR,PIK3C2A,PIK3C3,PIK3R1,PTE MHC_I N inos Hyper MAPK1 MHC_I inos Tumor JUN,IFNGR2,IRF1 MHC_I Estrogen-mediated S-phase SKP2 MHC_I Entry Estrogen-mediated S-phase Hyper E2F4 MHC_I
159 Entry Estrogen-mediated S-phase Entry GDNF Family Ligand- Receptor Interactions Tumor TFDP1,SKP2 MHC_I FGFR2 MHC_I Hyper MAPK1,ITPR2,PIK3R2 MHC_I Tumor JUN,PIK3C2A,PIK3C3,PIK3R1,ITPR3,RASA1 MHC_I GDNF Family Ligand- Receptor Interactions GDNF Family Ligand- Receptor Interactions Diphthamide Biosynthesis Tumor EEF2 MHC_I Thio-molybdenum Cofactor Tumor MOCOS MHC_I Biosynthesis 2-ketoglutarate Tumor DT,OGDH MHC_I Dehydrogenase Complex Palmitate Biosynthesis I Tumor FASN MHC_I (Animals) TCA Cycle II (Eukaryotic) Tumor ACO2,DT,OGDH MHC_I Anandamide Degradation Tumor FAAH2 MHC_I Glycerol-3-phosphate Shuttle Tumor GPD2 MHC_I Fatty Acid Biosynthesis Tumor FASN MHC_I Initiation II Zymosterol Biosynthesis Tumor HSD17B7,LBR MHC_I Glutamate Biosynthesis II Tumor GLUD1 MHC_I Glutamate Degradation X Tumor GLUD1 MHC_I Role of p14/p19arf in Tumor FGFR2 MHC_I Suppression Role of p14/p19arf in Tumor Hyper PIK3R2 MHC_I Suppression Role of p14/p19arf in Tumor Suppression Tumor PIK3C2A,PIK3C3,PIK3R1 MHC_I PI3K/AKT Hyper TSC1,MAPK1,TSC2,PIK3R2,THEM4 MHC_I PI3K/AKT Tumor AKT2,MTOR,PIK3R1,ILK,CTNNB1,PTEN MHC_I Interferon Hyper MED14 MHC_I
160 Interferon Tumor IFNGR2,IFNAR2,IRF1 MHC_I IL STAT5A,FGFR2 MHC_I IL-2 Hyper STAT5A,MAPK1,PIK3R2 MHC_I IL-2 Tumor AKT2,JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I IL FGFR2 MHC_I IL-4 Hyper PIK3R2,NFATC4 MHC_I IL-4 Tumor AKT2,MTOR,PIK3C2A,PIK3C3,PIK3R1 MHC_I Neurotrophin/TRK FGFR2 MHC_I Neurotrophin/TRK Hyper MAPK1,PIK3R2 MHC_I Neurotrophin/TRK Tumor JUN,PIK3C2A,PIK3C3,PIK3R1 MHC_I Cancer Drug Resistance By Drug Efflux Cancer Drug Resistance By Drug Efflux Actin Nucleation by ARP- WASP Complex Actin Nucleation by ARP- WASP Complex Actin Nucleation by ARP- WASP Complex Hyper MAPK1,PIK3R2 MHC_I Tumor BRAF,AKT2,PIK3R1,PTEN MHC_I FAP ROCK2,ROCK1,ACTR2,PPP1R12C,SOS1,BAIAP2, NCK1 Hyper ROCK1,NCK2,RHOQ Tumor ROCK1,SOS2,VASP
161 Estrogen Receptor FAP MED13,CDK8,MED20,MED15,SOS1,MED21,TBP,G TF2H1,NCOR1,MED13L,MAP2K1 Estrogen Receptor Estrogen Receptor Hyper MED23,MED MAPK1,TRRAP,CREBBP,NCOA1,NCOR2,SMARCA 4,MED14 Estrogen Receptor Tumor POLR2C,MED23,POLR2A,MAPK1,ERCC3,TRRAP, SOS2,NCOR1,PCK1,MED24,TAF2,SMARCA4 RhoA FAP ROCK2,ROCK1,ARHGAP5,ACTR2,LPAR6,BAIAP2, LPAR5,ARHGAP12,MYLK,ARHGAP1 RhoA RhoA Hyper ARHGEF1,PLD PTK2,ROCK1,ARHGAP5,ANLN,LPAR5,ARHGAP1, PI4KA,PKN1 RhoA Tumor ROCK1,PIKFYVE,RAPGEF6,ARHGEF1 Cell Cycle Control of Chromosomal Replication Cell Cycle Control of Chromosomal Replication Cell Cycle Control of Chromosomal Replication Cell Cycle Control of Chromosomal Replication FAP TOP2B,CDK12,CDK8,PRIM1,TOP2A,ORC CHEK2 Hyper LIG1,MCM2,CDK11B,POLD1 Tumor TOP2B,CDK12,ORC3,POLE,CDK4,MCM4,MCM7
162 Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis FAP IL6ST,PIK3C2B,FRZB,PDIA3,CSNK1A1,STAT3,RO CK1,ROCK2,CALM1 (includes others),plcb4,camk2d,fzd6,prkd3,map2k1,tc F7L2,LRP1,PPP3CA Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis Hyper PLCD3,FGFR2,TLR3,STAT ROCK1,PLCD3,NLK,MAPK1,TNFRSF1A,PRKCD,C REBBP,DVL1,PLCB3,PRKCH,CEBPB,PIK3R2,LRP1 Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis Tumor ROCK1,AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C 3,DVL1,CTNNB1,PLCH1,LRP1 Role of BRCA1 in DNA Damage Response FAP ABRAXAS1,TOPBP1,RFC2,ATR,SMARCD1,SMAR CC1,NBN Role of BRCA1 in DNA Damage Response Role of BRCA1 in DNA Damage Response Hyper PBRM1,TOPBP1,CHEK E2F4,FANCD2,SMARCB1,MSH6,HLTF,SMARCA4, NBN Role of BRCA1 in DNA Damage Response Tumor PBRM1,RBL2,MSH6,BABAM1,ARID2,SMARCA4,ML H1,RFC3 Wnt/β-catenin FAP AXIN2,FRZB,APPL2,FZD6,CSNK1A1,NR5A2,TLE1, BMPR2,SOX9,TCF7L2,LRP1
163 Wnt/β-catenin Hyper CSNK1E,NLK,TGFBR1,CSNK1G2,CSNK1G3,DVL1, CREBBP,MARK2,ACVR1,DVL3,LRP1 Wnt/β-catenin Tumor AKT2,JUN,HDAC1,DVL1,ILK,CTNNB1,LRP1 UDP-N-acetyl-D-glucosamine Biosynthesis II Mouse Embryonic Stem Cell Pluripotency Mouse Embryonic Stem Cell Pluripotency Mouse Embryonic Stem Cell Pluripotency Mouse Embryonic Stem Cell Pluripotency FAP GNPNAT1,PGM3 FAP IL6ST,PIK3C2B,SOS1,FZD6,BMPR2,STAT3,MAP2K 1,TCF7L BMP4,TYK2,FGFR2,STAT3 Hyper MAPK1,DVL1,CREBBP,DVL3,PIK3R2 Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,DVL1,SOS2,CTNN B1 Cell Cycle: G2/M DNA Damage Checkpoint Regulation Cell Cycle: G2/M DNA Damage Checkpoint Regulation Cell Cycle: G2/M DNA Damage Checkpoint Regulation FAP TOP2B,TRIP12,TOP2A,CCNB2,ATR CHEK2,SKP2 Tumor TOP2B,SKP2 Valine Degradation I FAP HIBCH,EHHADH,BCKDHB Melatonin FAP CALM1 (includes others),plcb4,camk2d,pdia3,map2k1,prkd3 Melatonin PLCD3
164 Melatonin Hyper PLCD3,MAPK1,PRKCD,PLCB3,PRKCH Melatonin Tumor BRAF,MAPK1,PLCH1 Oncostatin M FAP IL6ST,SOS1,STAT3,MAP2K1 Oncostatin M TYK2,STAT3 Oncostatin M Hyper STAT5A,MAPK1 Oncostatin M Tumor EPAS1,MAPK1 DNA damage-induced FAP RAD17,CCNB2,ATR 3σ DNA damage-induced Tumor AKT2 3σ GM-CSF FAP PIK3C2B,CAMK2D,SOS1,STAT3,MAP2K1,PPP3CA GM-CSF FGFR2,STAT3 GM-CSF Hyper ETS1,MAPK1,PIK3R2 GM-CSF Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,RACK1,SOS2 Transcriptional Regulatory Network in Embryonic Stem Cells Transcriptional Regulatory Network in Embryonic Stem Cells Transcriptional Regulatory Network in Embryonic Stem Cells Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency FAP TRIM24,RIF1,KAT6A,SKIL,STAT3 Hyper STAT KAT6A,CDX2 FAP IL6ST,PIK3C2B,RIF1,SOS1,FZD6,BMPR2,STAT3,M AP2K1
165 Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency BMP4,TYK2,FGFR2,STAT3 Tumor AKT2,RIF1,MAPK1,PIK3C2A,PIK3C3,DVL1,SOS2,C TNNB1 PPARα/RXRα Activation FAP CAND1,PLCB4,HELZ2,PDIA3,SOS1,PRKAA1,NCO A6,BMPR2,NCOR1,MAP2K1 PPARα/RXRα Activation PPARα/RXRα Activation Hyper PLCD3,MED23,CPT1B TGFBR1,PRKAB2,MAPK1,ADCY3,CREBBP,ACVR1,ADCY6,CKAP5,NFKB2,PLCD3,HELZ2,FASN,PLCB 3,NCOR2 PPARα/RXRα Activation Tumor SMAD2,MED23,MAPK1,SOS2,ADIPOR1,PLCH1,CA ND1,JUN,NFKBIA,GPD2,FASN,NCOR1,GOT2,MED 24 CNTF FAP IL6ST,PIK3C2B,SOS1,STAT3,MAP2K1 CNTF TYK2,FGFR2,STAT3 CNTF Tumor MTOR,MAPK1,PIK3C2A,PIK3C3,RPS6KB2 CCR3 in Eosinophils FAP ROCK2,ROCK1,PIK3C2B,CALM1 (includes others),plcb4,mylk,map2k1,prkd3 CCR3 in Eosinophils Hyper ROCK1,MAPK1,ITPR2,PRKCD,PLCB3,PRKCH,PIK 3R2
166 CCR3 in Eosinophils Tumor ROCK1,MAPK1,PIK3C2A,PIK3C3,RACK1,ITPR3 IL-3 FAP PIK3C2B,SOS1,STAT3,MAP2K1,PRKD3,PPP3CA IL-3 IL-3 Hyper FGFR2,STAT RAPGEF1,STAT5A,FOXO1,MAPK1,PRKCD,PRKCH,PIK3R2 IL-3 Tumor STAT6,AKT2,JUN,MAPK1,PIK3C2A,PIK3C3 Thrombin FAP ROCK2,ROCK1,PIK3C2B,PLCB4,CAMK2D,PDIA3,S OS1,TBP,MYLK,MAP2K1,PRKD3 Thrombin Thrombin Hyper PLCD3,FGFR2,ARHGEF1,ARHGEF CAMK1,MAPK1,ITPR2,ADCY3,ADCY6,NFKB2,ROC K1,PTK2,PLCD3,ARHGEF5,RHOQ,PRKCD,PLCB3, PRKCH,PIK3R2 Thrombin Tumor ROCK1,AKT2,MAPK1,PIK3C2A,PIK3C3,RACK1,ITP R3,ARHGEF1,PLCH1 Leptin in Obesity FAP PIK3C2B,PLCB4,PDIA3,PDE3B,STAT3,MAP2K1 Leptin in Obesity PLCD3,FGFR2,STAT3
167 Leptin in Obesity Hyper PLCD3,FOXO1,MAPK1,ADCY3,ADCY6,PLCB3,PIK 3R2 Leptin in Obesity Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,PLCH1 Assembly of RNA Polymerase FAP POLR1B,TBP I Complex UDP-N-acetyl-Dgalactosamine FAP GNPNAT1,PGM3 Biosynthesis II Thrombopoietin FAP PIK3C2B,SOS1,STAT3,MAP2K1,PRKD3 Thrombopoietin FGFR2,STAT3 Thrombopoietin Hyper STAT5A,MAPK1,PRKCD,PRKCH,PIK3R2 Thrombopoietin Tumor JUN,MAPK1,PIK3C2A,PIK3C3 Gαq FAP ROCK2,ROCK1,BTK,PIK3C2B,CALM1 (includes others),plcb4,map2k1,prkd3,ppp3ca Gαq Gαq Hyper FGFR2,PLD ROCK1,RHOQ,MAPK1,ITPR2,PRKCD,PLCB3,PRK CH,PIK3R2,NFKB2 Gαq Tumor ROCK1,AKT2,NFKBIA,PLD3,MAPK1,PIK3C2A,PIK3 C3,RACK1,ITPR3 Cleavage and Polyadenylation of Pre-mRNA Cleavage and Polyadenylation of Pre-mRNA FAP CPSF6,CSTF CPSF2
168 Cleavage and Polyadenylation Hyper CPSF1 of Pre-mRNA Glycogen Degradation II FAP PGM3,PYGL Glycogen Degradation II Hyper PGM2L1 Chemokine FAP ROCK2,CALM1 (includes others),plcb4,camk2d,map2k1 Chemokine Hyper PTK2,CAMK1,MAPK1,PLCB3 Chemokine Tumor JUN,MAPK1 RAR Activation FAP TRIM24,SDR16C5,RDH14,GTF2H1,NCOR1,SMAR CD1,SMARCC1,MAP2K1,RDH13,PRKD3 RAR Activation Hyper STAT5A,MAPK1,CREBBP,ADCY3,ADCY6,SMAD6, NFKB2,PSMC5,SMARCA4,PRKCD,SMARCB1,NCO A1,PRKCH,NCOR2,PIK3R2,RXRB,HLTF RAR Activation Tumor PBRM1,SMAD2,AKT2,JUN,MAPK1,ERCC3,NCOR1, ARID2,SMARCA4,PTEN Acute Myeloid Leukemia Acute Myeloid Leukemia Acute Myeloid Leukemia FAP PIK3C2B,SOS1,STAT3,MAP2K1,SPI1,TCF7L2 Hyper FGFR2,STAT STAT5A,MAPK1,PIK3R2,NFKB2
169 Acute Myeloid Leukemia Role of NFAT in Cardiac Hypertrophy Tumor BRAF,AKT2,MTOR,MAPK1,PIK3C2A,PIK3C3,SOS2,RPS6KB2 FAP IL6ST,PIK3C2B,CALM1 (includes others),plcb4,camk2d,pdia3,sos1,csnk1a1,m AP2K1,PRKD3,PPP3CA Role of NFAT in Cardiac Hypertrophy Hyper PLCD3,TGFBR1,CAMK1,MAPK1,HDAC8,ITPR2,PR KCD,ADCY3,ADCY6,PLCB3,PRKCH,PIK3R2,HDAC 5 Role of NFAT in Cardiac Hypertrophy Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,HDAC1,RACK1,ITP R3,SOS2,PLCH1,CABIN1 Aldosterone in Epithelial Cells FAP PIK3C2B,PLCB4,PDIA3,SLC12A2,HSPH1,SOS1,DN AJC12,MAP2K1,PRKD3 Aldosterone in Epithelial Cells Aldosterone in Epithelial Cells Hyper PLCD3,DNAJC13,FGFR PLCD3,MAPK1,DNAJB4,ITPR2,PRKCD,DNAJC12,H SPB11,PLCB3,PRKCH,NR3C2,DNAJB6,PIK3R2,PI4 KA Aldosterone in Epithelial Cells Tumor MAPK1,PIK3C2A,PIK3C3,ITPR3,SOS2,PIKFYVE,PL CH1
170 Protein Kinase A FAP PTPRD,PDIA3,AKAP9,MYLK,PYGL,ROCK1,ROCK2,CALM1 (includes others),plcb4,camk2d,pde3b,dusp11,pgp,prk D3,MAP2K1,TCF7L2,PPP3CA Protein Kinase A Protein Kinase A Hyper PLCD3,PTPN2,DUSP11,PTPRR FLNB,TGFBR1,PHKG2,MAPK1,PTPN23,ITPR2,CR EBBP,ADCY3,ADCY6,PTPN3,NFKB2,ROCK1,PTK2,PLCD3,PRKCD,PLCB3,PRKCH,PTPN21,DUSP16 Protein Kinase A Tumor ANAPC2,FLNB,PTPRK,MAPK1,AKAP9,RACK1,PTP RF,PLCH1,PTEN,ROCK1,HIST1H1B,BRAF,PHKB,N FKBIA,ITPR3,PTPN1,ANAPC5,CTNNB1,VASP Sumoylation Pathway FAP RANBP2,DAXX,SENP6,RFC2,SENP3,SP100 Sumoylation Pathway Hyper ETS1,TDG,RHOQ,SIRT1,CREBBP,NFKB2 Sumoylation Pathway Tumor RNF4,SAE1,NFKBIA,JUN,HDAC1,RFC3 ATM FAP HERC2,TOPBP1,RAD17,CCNB2,ATR,NBN ATM TOPBP1,CHEK2
171 ATM Hyper SMC3,HP1BP3,RNF168,FANCD2,TRRAP,CREBBP, TLK2,NBN ATM Tumor HERC2,NFKBIA,JUN,SMC2,TRRAP PI3K in B Lymphocytes FAP BTK,CALM1 (includes others),plcb4,camk2d,pdia3,map2k1,ppp3ca PI3K in B Lymphocytes PI3K in B Lymphocytes Hyper PLCD3,MAPK1,ITPR2,PLCB3,PIK3R2,NFKB2 Tumor AKT2,NFKBIA,JUN,MAPK1,ITPR3,PLCH1,PTEN Protein Ubiquitination Pathway FAP UBE2A,UBE2Q1,MED20,USP15,UBE3B,HSPH1,DN AJC12,USP54,USP30,USP33,USP34,UBE3A Protein Ubiquitination Pathway PAN2,PSMD10,USP5,UCHL5,DNAJC13,AMFR,SKP 2 Protein Ubiquitination Pathway Hyper USP14,DNAJB4,UBR2,DNAJC12,BIRC6,THOP1,PS MC5,USP48,PSMD11,HSPB11,DNAJB6,USP34,BIR C2 Protein Ubiquitination Pathway Tumor ANAPC2,USP15,USP5,HLA- A,USP36,USP9X,BIRC6,USP54,USP48,SKP2,USP3 2,PSMD12,ANAPC5,SMURF2,UBA1 p70s6k FAP BTK,PIK3C2B,PLCB4,PDIA3,SOS1,MAP2K1,PRKD 3
172 p70s6k p70s6k Hyper PLCD3,FGFR2,PLD PLCD3,MAPK1,PRKCD,PLCB3,PRKCH,PIK3R2,BC AP31 p70s6k Tumor AKT2,MTOR,MAPK1,PIK3C2A,EEF2,PIK3C3,SOS2, PLCH1 FLT3 in Hematopoietic Progenitor Cells FLT3 in Hematopoietic Progenitor Cells FLT3 in Hematopoietic Progenitor Cells FLT3 in Hematopoietic Progenitor Cells FAP PIK3C2B,SOS1,STAT3,MAP2K1,EIF4E Hyper FGFR2,STAT STAT5A,MAPK1,CREBBP,PIK3R2 Tumor STAT6,AKT2,MTOR,MAPK1,PIK3C2A,PIK3C3,SOS 2 Prolactin FAP PIK3C2B,SOS1,STAT3,MAP2K1,PRKD3 Prolactin Prolactin Hyper FGFR2,STAT STAT5A,MAPK1,PRKCD,CREBBP,PRKCH,CEBPB, PIK3R2 Prolactin Tumor JUN,MAPK1,PIK3C2A,PIK3C3,SOS2,IRF1 JAK/Stat FAP PIK3C2B,PIAS2,SOS1,STAT3,MAP2K1 JAK/Stat TYK2,FGFR2,STAT3
173 JAK/Stat Hyper STAT5A,MAPK1,CEBPB,PIK3R2,NFKB2 JAK/Stat Tumor STAT6,AKT2,MTOR,JUN,MAPK1,PIK3C2A,PIK3C3, PTPN1,SOS2 T Cell Receptor FAP BTK,PIK3C2B,CALM1 (includes others),sos1,map2k1,ppp3ca T Cell Receptor Tumor NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2,RASA 1 trna Charging FAP EARS2,IARS,FARSB trna Charging VARS trna Charging Hyper YARS,VARS2,TARS,SARS2,IARS trna Charging Tumor WARS,TARS2,EPRS Gα12/13 FAP ROCK2,ROCK1,BTK,PIK3C2B,LPAR6,LPAR5,MAP 2K1 Gα12/ FGFR2,ARHGEF1 Gα12/13 Hyper PTK2,ROCK1,MAPK1,LPAR5,PIK3R2,NFKB2 Gα12/13 Tumor ROCK1,AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C 3,ARHGEF1,CTNNB1,RASA1 Gap Junction FAP PIK3C2B,PLCB4,PDIA3,TJP1,SOS1,CSNK1A1,MAP 2K1,PRKD3,PPP3CA Gap Junction PLCD3,FGFR2
174 Gap Junction Hyper TUBB3,MAPK1,ITPR2,CSNK1G2,CSNK1G3,TUBG1,ADCY3,ADCY6,CSNK1E,PLCD3,TUBB6,PRKCD,P LCB3,PRKCH,PIK3R2 Gap Junction Tumor AKT2,TUBB3,MAPK1,PIK3C2A,TUBB4B,PIK3C3,IT PR3,SOS2,CTNNB1,NPR2,PLCH1 Glioma FAP PIK3C2B,CALM1 (includes others),camk2d,sos1,map2k1,prkd3 Glioma Hyper E2F4,CAMK1,MAPK1,PRKCD,CDKN2C,PRKCH,PIK 3R2 Glioma Tumor AKT2,RBL2,MTOR,TFDP1,MAPK1,PIK3C2A,PIK3C 3,SOS2,CDK4,PTEN Pyridoxal 5'-phosphate Salvage Pathway Pyridoxal 5'-phosphate Salvage Pathway FAP CDK8,PRKAA1,CSNK1A1,MAP2K1 Hyper PRKX,DMPK,MAPK1,PRKCD,GRK6,PRKCH,PKN1 Pyridoxal 5'-phosphate Tumor BRAF,AKT2,DMPK,MAPK1,CDK4,NEK2 Salvage Pathway HGF FAP PIK3C2B,ELF3,SOS1,STAT3,MAP2K1,PRKD3 HGF ELF4,FGFR2,STAT3
175 HGF Hyper PTK2,ETS1,RAPGEF1,MAP3K11,MAPK1,PRKCD,P RKCH,PIK3R2 HGF Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 UVB-Induced MAPK FAP PIK3C2B,MAP2K1,PRKD3,EIF4E UVB-Induced MAPK FGFR2 UVB-Induced MAPK Hyper MAPK1,PRKCD,PRKCH,PIK3R2 UVB-Induced MAPK Tumor MTOR,JUN,MAPK1,PIK3C2A,PIK3C3 ErbB2-ErbB3 FAP PIK3C2B,SOS1,STAT3,MAP2K1 ErbB2-ErbB TYK2,FGFR2,STAT3 ErbB2-ErbB3 Hyper STAT5A,FOXO1,MAPK1,ERBB2,PIK3R2 ErbB2-ErbB3 Tumor JUN,MAPK1,PIK3C2A,PIK3C3,SOS2,PTEN EGF FAP PIK3C2B,SOS1,STAT3,MAP2K1 EGF FGFR2,STAT3 EGF Hyper MAPK1,ITPR2,PIK3R2 EGF Tumor AKT2,MTOR,JUN,MAPK1,PIK3C2A,PIK3C3,ITPR3, SOS2,RASA1
176 Breast Cancer Regulation by Stathmin1 FAP ROCK2,ROCK1,PIK3C2B,CALM1 (includes others),plcb4,camk2d,sos1,map2k1,prkd3 Breast Cancer Regulation by Stathmin1 Breast Cancer Regulation by Stathmin1 Hyper FGFR2,ARHGEF1,ARHGEF TUBB3,E2F4,CAMK1,MAPK1,ITPR2,TUBG1,ADCY3,ADCY6,ROCK1,ARHGEF5,TUBB6,PRKCD,PLCB3, PRKCH,PIK3R2 Breast Cancer Regulation by Stathmin1 Tumor ROCK1,TUBB3,MAPK1,PIK3C2A,TUBB4B,PIK3C3, ARHGEF7,RACK1,ITPR3,SOS2,ARHGEF1 ErbB FAP PIK3C2B,SOS1,NCK1,MAP2K1,PRKD3 ErbB ErbB Hyper FGFR NCK2,FOXO1,MAPK1,PRKCD,PRKCH,ERBB2,PIK3 R2 ErbB Tumor MTOR,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 TR/RXR Activation FAP PIK3C2B,PDE3B,NCOA6,NCOR1,THRA TR/RXR Activation Hyper SREBF1,FASN,NCOA1,STRBP,PIK3R2,NCOR2,RX RB
177 TR/RXR Activation Tumor SLC16A3,AKT2,MTOR,PIK3C2A,SREBF1,PIK3C3,F ASN,NCOR1,PCK1 Axonal Guidance FAP ACTR2,PIK3C2B,PDIA3,NCK1,EIF4E,ROCK1,HER C2,ROCK2,PLCB4,MICAL1,SOS1,BAIAP2,MKNK1, FZD6,PRKD3,MAP2K1,PPP3CA Axonal Guidance Hyper TUBB3,ADAM17,MAPK1,NTN4,TUBG1,SEMA6A,PL XNA2,GIT1,RAC3,ROCK1,NCK2,PTK2,PLCD3,TUB B6,PRKCD,PLCB3,EPHB3,PRKCH,PLXNB2,ERBB2,PIK3R2,EPHA2 Axonal Guidance Tumor TUBB3,AKT2,MAPK1,PIK3C2A,TUBB4B,ARHGEF7, SOS2,RACK1,PLCH1,ROCK1,HERC2,PIK3C3,PLX NB1,LNPEP,CHMP1A,RASA1,EPHA2,VASP,FARP2 Role of NFAT in Regulation of the Immune Response FAP BTK,PIK3C2B,CALM1 (includes others),plcb4,sos1,csnk1a1,map2k1,ppp3ca Role of NFAT in Regulation of the Immune Response Hyper CSNK1E,MAPK1,CSNK1G2,ITPR2,CSNK1G3,PLCB 3,PIK3R2,NFKB2
178 Role of NFAT in Regulation of the Immune Response Tumor AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,HLA- A,PIK3C3,RACK1,ITPR3,SOS2,CABIN1 CREB in Neurons FAP PIK3C2B,CALM1 (includes others),plcb4,camk2d,pdia3,sos1,tbp,map2k1,prkd3 CREB in Neurons Hyper PLCD3,MAPK1,ITPR2,PRKCD,CREBBP,ADCY3,AD CY6,PLCB3,PRKCH,PIK3R2 CREB in Neurons Tumor AKT2,POLR2C,POLR2A,MAPK1,PIK3C2A,PIK3C3, RACK1,ITPR3,SOS2,PLCH1 Role of JAK family kinases in IL-6-type Cytokine Role of JAK family kinases in IL-6-type Cytokine FAP IL6ST,STAT TYK2,STAT3 Hyper STAT5A,MAPK1 Tumor MAPK1 Role of JAK family kinases in IL-6-type Cytokine Role of JAK family kinases in IL-6-type Cytokine VEGF FAP ROCK2,ROCK1,PIK3C2B,SOS1,MAP2K1 VEGF Hyper PTK2,ROCK1,FOXO1,MAPK1,PIK3R2,ACTN1 VEGF Tumor ROCK1,AKT2,MAPK1,PIK3C2A,PIK3C3,SOS2 EIF2 FAP PIK3C2B,PTBP1,SOS1,EIF5B,EIF4G3,RPS17,EIF2 AK3,MAP2K1,EIF4E
179 EIF2 Tumor EIF2AK1,EIF2AK4,PTBP1,AKT2,WARS,MAPK1,PIK 3C2A,SREBF1,PIK3C3,EIF5B,SOS2,EIF4G1 Integrin FAP ROCK1,ARHGAP5,PIK3C2B,ACTR2,SOS1,MYLK,IT GA1,NCK1,MAP2K1 Integrin Hyper PTK2,ROCK1,NCK2,ARHGAP5,RAPGEF1,RHOQ,M AP3K11,MAPK1,PIK3R2,GIT1,RAC3,ACTN1 Integrin Tumor AKT2,MAPK1,PIK3C2A,ARHGEF7,SOS2,PIKFYVE,I LK,TLN1,PTEN,ROCK1,BRAF,PARVB,PIK3C3,ZYX, ITGA1,VASP Assembly of RNA Polymerase II Complex Assembly of RNA Polymerase II Complex GDNF Family Ligand- Receptor Interactions GDNF Family Ligand- Receptor Interactions GDNF Family Ligand- Receptor Interactions GDNF Family Ligand- Receptor Interactions Superpathway of Cholesterol Biosynthesis Superpathway of Cholesterol Biosynthesis FAP TBP,GTF2H1,GTF2E2 Tumor POLR2C,POLR2A,ERCC3,TAF2 FAP PIK3C2B,SOS1,NCK1,MAP2K FGFR2 Hyper MAPK1,ITPR2,PIK3R2 Tumor JUN,MAPK1,PIK3C2A,PIK3C3,ITPR3,SOS2,RASA1 FAP DHCR7,ACAT2 Hyper LBR
180 Superpathway of Cholesterol Biosynthesis Non-Small Cell Lung Cancer Non-Small Cell Lung Cancer Non-Small Cell Lung Cancer Non-Small Cell Lung Cancer Tumor PMVK,HSD17B7,HMGCR,LBR FAP STK4,PIK3C2B,SOS1,MAP2K FGFR2 Hyper MAPK1,ITPR2,ERBB2,PIK3R2,RXRB Tumor AKT2,TFDP1,MAPK1,PIK3C2A,PIK3C3,ITPR3,SOS 2,CDK4 Adipogenesis pathway FAP ATG5,KAT6A,FZD6,GTF2H1,BMPR2,SOX9 Adipogenesis pathway Adipogenesis pathway Hyper BMP4,FGFR FOXO1,HDAC8,TNFRSF1A,SREBF1,KAT6A,SIRT1, SAP130,CEBPB,KAT2A,HDAC5 Adipogenesis pathway Tumor SREBF1,ERCC3,HDAC1,CTNNB1 Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes FAP CALM1 (includes others),sos1,map2k1,ppp3ca Hyper TGFBR1,MAPK1,NFKB2 Tumor SMAD2,NFKBIA,JUN,MAPK1,SOS2 Telomerase FAP PIK3C2B,ELF3,SOS1,TEP1,MAP2K1 Telomerase ELF4,FGFR2
181 Telomerase Hyper ETS1,MAPK1,HDAC8,TEP1,PIK3R2,POT1,HDAC5 Telomerase Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,HDAC1,SOS2 Erythropoietin FAP PIK3C2B,SOS1,MAP2K1,PRKD3 Erythropoietin Erythropoietin Hyper FGFR STAT5A,MAPK1,PRKCD,PRKCH,PIK3R2,NFKB2 Erythropoietin Tumor AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 Insulin Receptor FAP PIK3C2B,PDE3B,SOS1,NCK1,MAP2K1,EIF4E Insulin Receptor Hyper TSC1,RAPGEF1,RHOQ,FOXO1,MAPK1,TSC2,PIK3 R2 Insulin Receptor Tumor AKT2,MTOR,MAPK1,PIK3C2A,PIK3C3,PTPN1,SOS 2,RPS6KB2,STX4,PTPRF,PTEN Tight Junction FAP CPSF6,EPB41,TJP1,AFDN,MYLK,GOSR1,CSTF3 Tight Junction Hyper EPB41,MYH9,TGFBR1,TNFRSF1A,NECTIN3,MARK 2,NAPG,CPSF1,CASK,NFKB2,PATJ,NECTIN2 Tight Junction Tumor AKT2,JUN,NECTIN3,CDK4,SYMPK,STX4,CTNNB1, VASP,PTEN
182 Hereditary Breast Cancer FAP PIK3C2B,RFC2,ATR,SMARCD1,SMARCC1,NBN Hereditary Breast Cancer Hereditary Breast Cancer Hyper PBRM1,FGFR2,CHEK HDAC8,FANCD2,SMARCB1,XPC,CREBBP,TUBG1, MSH6,PIK3R2,HLTF,SMARCA4,NBN,HDAC5 Hereditary Breast Cancer Tumor PBRM1,AKT2,POLR2C,POLR2A,PIK3C2A,PIK3C3, HDAC1,MSH6,CDK4,ARID2,SMARCA4,MLH1,PTEN,RFC3 Epithelial Adherens Junction Epithelial Adherens Junction FAP ACTR2,BAIAP2,AFDN,BMPR2,TCF7L2,CLIP1 Hyper RAPGEF1,TUBB3,TGFBR1,MYH9,TUBB6,SORBS1, KEAP1,NECTIN3,TUBG1,ACVR1,CLIP1,ACTN1,NE CTIN2 Epithelial Adherens Junction Tumor AKT2,TUBB3,TUBB4B,NECTIN3,ZYX,CLINT1,CTN NB1,NOTCH1,FARP2,CTNND1,PTEN Molecular Mechanisms of Cancer FAP CDK12,PIK3C2B,BMPR2,NBN,DAXX,PLCB4,CAMK 2D,CDK8,SOS1,FZD6,ATR,PRKD3,MAP2K1,LRP1 Molecular Mechanisms of Cancer ZBTB17,BMP4,TYK2,FGFR2,ARHGEF1,ARHGEF9, CHEK2
183 Molecular Mechanisms of Cancer Hyper RAPGEF1,E2F4,TGFBR1,MAPK1,DVL1,ADCY3,CR EBBP,ADCY6,SMAD6,CDKN2C,NFKB2,RAC3,NBN, PTK2,ARHGEF5,NLK,RHOQ,FANCD2,FOXO1,CDK 11B,PRKCD,PLCB3,PRKCH,PIK3R2,LRP1,BIRC2 Molecular Mechanisms of Cancer Tumor CDK12,SMAD2,AKT2,TFDP1,MAPK1,PIK3C2A,ARH GEF7,SOS2,DVL1,CDK4,APAF1,ARHGEF1,BRAF,J UN,NFKBIA,PIK3C3,CTNNB1,RASA1,NOTCH1,LRP 1,CTNND1 NF-κB Activation by Viruses FAP PIK3C2B,TBP,ITGA1,PRKD3 NF-κB Activation by Viruses FGFR2 NF-κB Activation by Viruses Hyper MAPK1,PRKCD,PRKCH,PIK3R2,NFKB2 NF-κB Activation by Viruses Tumor AKT2,NFKBIA,MAPK1,PIK3C2A,PIK3C3,ITGA1 PEDF FAP ROCK2,ROCK1,PIK3C2B,TCF7L2 PEDF PEDF Hyper FGFR ROCK1,HNF1B,WASF2,MAPK1,PIK3R2,NFKB2,PN PLA2
184 PEDF Tumor ROCK1,AKT2,NFKBIA,MAPK1,PIK3C2A,PIK3C3 PDGF FAP PIK3C2B,SOS1,STAT3,MAP2K1 PDGF TYK2,FGFR2,STAT3 PDGF Tumor JUN,MAPK1,PIK3C2A,PIK3C3,SOS2,RASA1 NGF FAP ROCK2,ROCK1,PIK3C2B,SOS1,MAP2K1 NGF NGF Hyper FGFR2,SMPD ROCK1,MAP3K11,MAPK1,PRKCD,CREBBP,TRIO, PIK3R2,NFKB2 NGF Tumor ROCK1,AKT2,MAPK1,PIK3C2A,PIK3C3,SOS2,RPS 6KB2,TRIO Renin-Angiotensin FAP PIK3C2B,SOS1,STAT3,MAP2K1,PRKD3 Renin-Angiotensin Renin-Angiotensin Hyper FGFR2,STAT PTK2,MAPK1,ITPR2,PRKCD,ADCY3,ADCY6,PRKC H,PIK3R2,NFKB2 Renin-Angiotensin Tumor JUN,MAPK1,PIK3C2A,PIK3C3,ITPR3,SOS2 Glucocorticoid Receptor FAP PIK3C2B,SOS1,PRKAA1,TBP,GTF2H1,GTF2E2,NC OR1,SMARCD1,SMARCC1,STAT3,MAP2K1,PPP3C A
185 Glucocorticoid Receptor Hyper STAT5A,TGFBR1,PRKAB2,MAPK1,CREBBP,GTF2 E2,CEBPB,SMARCA4,MED14,SMARCB1,NCOA1,N R3C2,NCOR2,PIK3R2,HLTF Glucocorticoid Receptor Tumor PBRM1,SMAD2,AKT2,MAPK1,PIK3C2A,SOS2,ARID 2,PCK1,SMARCA4,JUN,POLR2A,POLR2C,NFKBIA, ERCC3,PIK3C3,NCOR1,TAF2 Endometrial Cancer FAP PIK3C2B,SOS1,MAP2K1 Endometrial Cancer FGFR2 Endometrial Cancer Hyper MAPK1,ERBB2,PIK3R2 Endometrial Cancer Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,SOS2,ILK,CTNNB1, MLH1,PTEN IL-2 FAP PIK3C2B,SOS1,MAP2K1 IL FGFR2 IL-2 Hyper STAT5A,MAPK1,PIK3R2 IL-2 Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 Salvage Pathways of Pyrimidine Ribonucleotides Salvage Pathways of Pyrimidine Ribonucleotides FAP CDK8,PRKAA1,CSNK1A1,MAP2K1 Hyper PRKX,DMPK,MAPK1,PRKCD,GRK6,PRKCH,UCKL1,PKN1
186 Salvage Pathways of Tumor BRAF,AKT2,DMPK,MAPK1,CDK4,PCK1,NEK2 Pyrimidine Ribonucleotides Mevalonate Pathway I FAP ACAT2 Mevalonate Pathway I Tumor PMVK,HMGCR Endothelin-1 FAP PNPLA8,PIK3C2B,PLCB4,ABHD3,PDIA3,SOS1,PR KD3 Endothelin-1 Endothelin-1 Hyper PLCD3,FGFR2,PLD PLCD3,MAPK1,ITPR2,CASP2,PRKCD,ADCY3,ADC Y6,PLCB3,PRKCH,PIK3R2 Endothelin-1 Tumor PLBD1,BRAF,PLD3,JUN,MAPK1,PIK3C2A,PIK3C3,I TPR3,PLCH1,CASP5 Regulation of the Epithelial- Mesenchymal Transition Pathway Regulation of the Epithelial- Mesenchymal Transition Pathway Regulation of the Epithelial- Mesenchymal Transition Pathway Regulation of the Epithelial- Mesenchymal Transition Pathway FAP PIK3C2B,ESRP2,SOS1,FZD6,STAT3,MAP2K1,TCF 7L2 Hyper TYK2,FGFR2,STAT ETS1,MAML1,ADAM17,TGFBR1,MAPK1,DVL1,DVL 3,PIK3R2,NFKB2 Tumor BRAF,SMAD2,AKT2,MAPK1,PIK3C2A,PIK3C3,DVL 1,SOS2,CTNNB1,NOTCH1 B Cell Receptor FAP BTK,PIK3C2B,CALM1 (includes others),camk2d,sos1,map2k1,ppp3ca
187 B Cell Receptor Hyper PTK2,ETS1,MAP3K11,FOXO1,MAPK1,CREBBP,PI K3R2,NFKB2 B Cell Receptor Tumor AKT2,MTOR,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2,RPS6KB2,PTEN CD28 in T Helper Cells FAP PIK3C2B,CALM1 (includes others),actr2,map2k1,ppp3ca CD28 in T Helper Cells Tumor AKT2,NFKBIA,JUN,PIK3C2A,HLA-A,PIK3C3,ITPR3 Cholesterol Biosynthesis I FAP DHCR7 Cholesterol Biosynthesis I Hyper LBR Cholesterol Biosynthesis I Tumor HSD17B7,LBR Cholesterol Biosynthesis II FAP DHCR7 (via 24,25-dihydrolanosterol) Cholesterol Biosynthesis II Hyper LBR (via 24,25-dihydrolanosterol) Cholesterol Biosynthesis II Tumor HSD17B7,LBR (via 24,25-dihydrolanosterol) Cholesterol Biosynthesis III FAP DHCR7 (via Desmosterol) Cholesterol Biosynthesis III Hyper LBR (via Desmosterol) Cholesterol Biosynthesis III Tumor HSD17B7,LBR (via Desmosterol) IL-6 FAP IL6ST,PIK3C2B,SOS1,STAT3,MAP2K1 IL-6 IL-6 Hyper FGFR2,STAT MAPK1,TNFRSF1A,CEBPB,PIK3R2,NFKB2
188 IL-6 Tumor AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 Actin Cytoskeleton FAP ROCK2,ROCK1,PIK3C2B,ACTR2,SOS1,BAIAP2,MY LK,MAP2K1 Actin Cytoskeleton Actin Cytoskeleton Hyper DIAPH2,FGFR2,ARHGEF PTK2,ROCK1,DIAPH1,MYH9,CYFIP2,WASF2,MAP K1,TRIO,PIK3R2,GIT1,RAC3,ACTN1 Actin Cytoskeleton Tumor ROCK1,MAPK1,PIK3C2A,PIK3C3,DIAPH3,ARHGEF 7,SOS2,PIKFYVE,TRIO,ARHGEF1,TLN1,NCKAP1 FAK FAP PIK3C2B,HMMR,SOS1,MAP2K1 FAK Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,ARHGEF7,SOS2,T LN1,PTEN Dopamine-DARPP32 Feedback in camp FAP CALM1 (includes others),plcb4,pdia3,csnk1a1,prkd3,ppp3ca Dopamine-DARPP32 Feedback in camp Hyper CSNK1E,PLCD3,CSNK1G2,ITPR2,CSNK1G3,PRKC D,CREBBP,ADCY3,ADCY6,PLCB3,ATP2A3,PRKCH mediated FAP PIK3C2B,PLCB4,PDIA3,MAP2K1,PRKD mediated PLCD3,FGFR2
189 mediated Hyper TSC1,TUBB3,MAPK1,TNFRSF1A,TUBG1,PLCD3,F OXO1,TUBB6,PRKCD,TSC2,PLCB3,PRKCH,PIK3R mediated Tumor AKT2,TUBB3,JUN,MAPK1,PIK3C2A,TUBB4B,PIK3C 3,PLCH1 Glioblastoma Multiforme Glioblastoma Multiforme Glioblastoma Multiforme FAP PIK3C2B,PLCB4,PDIA3,SOS1,FZD6,MAP2K1 Hyper PLCD3,FGFR PLCD3,TSC1,E2F4,RHOQ,FOXO1,MAPK1,ITPR2,P RKCD,TSC2,PLCB3,PIK3R2 Glioblastoma Multiforme Tumor AKT2,MTOR,MAPK1,PIK3C2A,PIK3C3,ITPR3,SOS2,CDK4,CTNNB1,PLCH1,PTEN RANK in Osteoclasts FAP PIK3C2B,CALM1 (includes others),map2k1,ppp3ca RANK in Osteoclasts RANK in Osteoclasts DNA Double-Strand Break Repair by Homologous Recombination DNA Double-Strand Break Repair by Homologous Recombination Hyper MAP3K11,MAPK1,PIK3R2,NFKB2,BIRC2 Tumor AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3 FAP NBN Hyper LIG1,ATRX,NBN DNA Double-Strand Break FAP NBN
190 Repair by Non-Homologous End Joining DNA Double-Strand Break Repair by Non-Homologous End Joining DNA Double-Strand Break Repair by Non-Homologous End Joining Hyper NBN Tumor XRCC6,LIG3 CXCR4 FAP ROCK2,ROCK1,PIK3C2B,PLCB4,MAP2K1,PRKD3 CXCR4 Hyper PTK2,ROCK1,RHOQ,MAPK1,ITPR2,PRKCD,ADCY 3,ADCY6,PLCB3,PRKCH,PIK3R2 CXCR4 Tumor ROCK1,AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,RACK 1,ITPR3 P2Y Purigenic Receptor Pathway P2Y Purigenic Receptor Pathway P2Y Purigenic Receptor Pathway FAP PIK3C2B,PLCB4,PDIA3,MAP2K1,PRKD3 Hyper PLCD3,FGFR PLCD3,MAPK1,PRKCD,CREBBP,ADCY3,ADCY6,P LCB3,PRKCH,PIK3R2,NFKB2 P2Y Purigenic Receptor Pathway Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,RACK1,PLCH1 Superpathway of Inositol Phosphate Compounds FAP PPFIA1,PIK3C2B,PLCB4,PPP1R12C,DUSP11,PPP 4R1,PPP3CA,SACM1L Superpathway of Inositol Phosphate Compounds PLCD3,IPMK,PTPN2,DUSP11,FGFR2
191 Superpathway of Inositol Phosphate Compounds Hyper MTMR6,PLCD3,PTPN23,SGPP2,PLCB3,PPM1F,ER BB2,PPP4R1,PIK3R2,PI4KA,DUSP16 Superpathway of Inositol Phosphate Compounds Tumor PIK3C2A,PIK3C3,PTPN1,PIKFYVE,RASA1,PMPCA, PLCH1,PTPRF,PPIP5K2,PTEN Androgen FAP CALM1 (includes others),tbp,gtf2h1,gtf2e2,prkd3 Androgen Hyper CALR,MAPK1,PRKCD,CREBBP,NCOA1,GTF2E2,P RKCH,NFKB2 Androgen Tumor POLR2C,POLR2A,JUN,MAPK1,ERCC3,RACK1,TAF 2 STAT3 Pathway FAP BMPR2,STAT3,MAP2K1 STAT3 Pathway STAT3 Pathway Chronic Myeloid Leukemia Chronic Myeloid Leukemia PTPN2,TYK2,FGFR2,STAT3 Hyper TGFBR1,MAP3K11,MAPK1 FAP PIK3C2B,SOS1,CBLC,MAP2K1 Hyper STAT5A,E2F4,TGFBR1,MAPK1,HDAC8,PIK3R2,NF KB2,HDAC5 Chronic Myeloid Leukemia Tumor AKT2,RBL2,TFDP1,MAPK1,PIK3C2A,PIK3C3,HDAC 1,SOS2,CDK4 UVA-Induced MAPK FAP PIK3C2B,PLCB4,PDIA3,PARP14 UVA-Induced MAPK PLCD3,FGFR2,SMPD1,PARP9
192 UVA-Induced MAPK Hyper PLCD3,MAPK1,PLCB3,PIK3R2 UVA-Induced MAPK Tumor MTOR,JUN,MAPK1,PIK3C2A,PIK3C3,RPS6KB2,PL CH1,PARP14 IGF-1 FAP PIK3C2B,SOS1,STAT3,MAP2K1 IGF FGFR2,STAT3 IGF-1 Hyper PTK2,FOXO1,MAPK1,PIK3R2 IGF-1 Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2,RPS6K B2,RASA1 Mismatch Repair in FAP RFC2 Eukaryotes Mismatch Repair in Hyper MSH6,POLD1 Eukaryotes Mismatch Repair in Tumor MSH6,MLH1,RFC3 Eukaryotes IL-9 FAP PIK3C2B,STAT3 IL FGFR2,STAT3 IL-9 Hyper STAT5A,PIK3R2,NFKB2 IL-9 Tumor PIK3C2A,PIK3C3 Phospholipase C FAP BTK,MARCKS,CALM1 (includes others),plcb4,sos1,map2k1,prkd3,ppp3ca Phospholipase C PLCD3,ARHGEF1,ARHGEF9,PLD1
193 Phospholipase C Hyper HDAC8,MAPK1,ITPR2,CREBBP,ADCY3,ADCY6,NF KB2,HDAC5,PLCD3,ARHGEF5,RHOQ,PRKCD,PLC B3,PRKCH Phospholipase C Tumor PLD3,MAPK1,ARHGEF7,HDAC1,RACK1,ITPR3,SO S2,ARHGEF1 IL-15 FAP PIK3C2B,STAT3,MAP2K1 IL TYK2,FGFR2,STAT3 IL-15 Hyper PTK2,STAT5A,MAPK1,PIK3R2,NFKB2,RAC3 IL-15 Tumor STAT6,AKT2,MAPK1,PIK3C2A,PIK3C3 Angiopoietin FAP PIK3C2B,SOS1,NCK1 Angiopoietin FGFR2 Angiopoietin Hyper PTK2,STAT5A,FOXO1,PIK3R2,NFKB2 Angiopoietin Tumor AKT2,NFKBIA,PIK3C2A,PIK3C3,RASA1 Neurotrophin/TRK FAP PIK3C2B,SOS1,MAP2K1 Neurotrophin/TRK FGFR2 Neurotrophin/TRK Hyper MAPK1,CREBBP,PIK3R2 Neurotrophin/TRK Tumor JUN,MAPK1,PIK3C2A,PIK3C3,SOS2
194 CD40 FAP PIK3C2B,STAT3,MAP2K1 CD FGFR2,STAT3 CD40 Hyper MAPK1,PIK3R2,NFKB2 CD40 Tumor NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3 IL-17A in Airway Cells IL-17A in Airway Cells IL-17A in Airway Cells IL-17A in Airway Cells FAP PIK3C2B,STAT3,MAP2K TYK2,FGFR2,STAT3 Hyper MAPK1,PIK3R2,NFKB2 Tumor AKT2,NFKBIA,MAPK1,PIK3C2A,PIK3C3,MUC5B,PT EN Paxillin FAP PIK3C2B,SOS1,ITGA1,NCK1 Paxillin Hyper PTK2,NCK2,MAPK1,PIK3R2,ACTN1 Paxillin Tumor MAPK1,PIK3C2A,PIK3C3,ARHGEF7,SOS2,TLN1,IT GA1 p53 FAP PIK3C2B,STAG1,TOPBP1,ATR p TOPBP1,FGFR2,CHEK2 p53 Tumor AKT2,JUN,PIK3C2A,PIK3C3,HDAC1,CCNK,APAF1, CDK4,CTNNB1,PTEN
195 Colorectal Cancer Metastasis FAP IL6ST,PIK3C2B,SOS1,FZD6,STAT3,MAP2K1,TCF7 L2,LRP1 Colorectal Cancer Metastasis Colorectal Cancer Metastasis Hyper TYK2,FGFR2,TLR3,STAT E2F4,RHOQ,TGFBR1,MAPK1,TNFRSF1A,DVL1,AD CY3,ADCY6,MSH6,PIK3R2,NFKB2,LRP1 Colorectal Cancer Metastasis Tumor SMAD2,AKT2,PIK3C2A,MAPK1,SOS2,RACK1,DVL1,MLH1,BRAF,JUN,PIK3C3,MSH6,CTNNB1,LRP1 Renal Cell Carcinoma Renal Cell Carcinoma Renal Cell Carcinoma Renal Cell Carcinoma Huntington's Disease FAP PIK3C2B,SOS1,MAP2K EGLN3,FGFR2 Hyper ETS1,RAPGEF1,MAPK1,CREBBP,PIK3R2 Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 FAP PIK3C2B,PLCB4,SOS1,TBP,PSME4,NCOR1,GOSR 1,PRKD3 Huntington's Disease Hyper SDHB,HDAC8,MAPK1,G,CREBBP,IFT57,NAPG, HDAC5,DNM1,CASP2,PRKCD,PSME4,PLCB3,PRK CH,NCOR2,PIK3R2
196 Huntington's Disease Tumor AKT2,MAPK1,PIK3C2A,HDAC1,SOS2,RACK1,APA F1,MTOR,JUN,POLR2A,POLR2C,PIK3C3,HTT,PSM E4,NCOR1,RASA1,CASP5,DNM2 Rac FAP PIK3C2B,ACTR2,BAIAP2,MAP2K1 Rac Rac Hyper FGFR2,PLD PTK2,MCF2L,CYFIP2,MAP3K11,MAPK1,PIK3R2,NF KB2,PI4KA Rac Tumor JUN,MAPK1,PIK3C2A,PIK3C3,PIKFYVE,NCKAP1 Sirtuin Pathway FAP ATG5,POLR1B,ATG4D,PRKAA1,SIRT3,STAT3,NDU FA1,TOMM70,NBN Sirtuin Pathway Sirtuin Pathway Hyper CPT1A,ATG9A,CPT1B,SIRT6,ZBTB14,G6PD,STAT PPID,SDHB,MAPK1,G,SF3A1,NFKB2,XPA,NBN, NDUFS1,FOXO1,ATG4D,SREBF1,SIRT1,XPC,NDU FS2,KAT2A Sirtuin Pathway Tumor TOMM40,EPAS1,TIMM44,MAPK1,SMARCA5,PCK1, PFKM,HIST1H1B,MTOR,NDUFS1,JUN,SREBF1,ND UFA11,XRCC6,GOT2
197 icos-icosl in T Helper Cells FAP PIK3C2B,CALM1 (includes others),camk2d,ppp3ca icos-icosl in T Helper Cells Tumor AKT2,NFKBIA,PIK3C2A,HLA- A,PIK3C3,ITPR3,PTEN Nur77 in T Lymphocytes Nur77 in T Lymphocytes FAP CALM1 (includes others),ppp3ca Tumor HLA-A,HDAC1,APAF1,CABIN1 Neuregulin FAP SOS1,MAP2K1,PRKD3 Neuregulin Hyper STAT5A,ADAM17,MAPK1,PRKCD,PRKCH,ERBB2, PIK3R2 Neuregulin Tumor AKT2,MTOR,MAPK1,SOS2,RPS6KB2,PTEN TGF-β FAP SOS1,BMPR2,MAP2K1 TGF-β BMP4 TGF-β Hyper TGFBR1,MAPK1,CREBBP,SKI,ACVR1,SMAD6 TGF-β Tumor ZFYVE9,SMAD2,JUN,MAPK1,HDAC1,SOS2,SMUR F2 HER-2 in Breast Cancer HER-2 in Breast Cancer HER-2 in Breast Cancer FAP PIK3C2B,SOS1,PRKD3 Hyper FGFR TSC1,FOXO1,PRKCD,TSC2,PRKCH,ERBB2,PIK3R 2
198 HER-2 in Breast Tumor AKT2,PIK3C2A,PIK3C3,SOS2 Cancer Melanoma FAP PIK3C2B,MAP2K1 Melanoma FGFR2 Melanoma Hyper MAPK1,PIK3R2 Melanoma Tumor BRAF,AKT2,MAPK1,PIK3C2A,PIK3C3,CDK4,PTEN Apoptosis FAP ROCK1,MAP2K1,AIFM1 Apoptosis Hyper ACIN1,ROCK1,MAPK1,TNFRSF1A,CASP2,BIRC6,N FKB2,AIFM1,BIRC2 Apoptosis Tumor ACIN1,ROCK1,NFKBIA,MAPK1,BIRC6,APAF1 IL-22 FAP STAT3 IL TYK2,STAT3 IL-22 Hyper STAT5A,MAPK1 IL-22 Tumor AKT2,MAPK1,IL10RB Role of JAK1, JAK2 and TYK2 FAP STAT3 in Interferon Role of JAK1, JAK2 and TYK PTPN2,TYK2,STAT3 in Interferon Role of JAK1, JAK2 and TYK2 Hyper NFKB2 in Interferon Role of JAK1, JAK2 and TYK2 Tumor IFNAR2 in Interferon CDP-diacylglycerol FAP TAZ Biosynthesis I CDP-diacylglycerol LPCAT4
199 Biosynthesis I CDP-diacylglycerol Biosynthesis I Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes Hyper TAMM41,LPCAT2,AGPAT3 FAP ACTR2,NCK1,PRKD3 Hyper PLD NCK2,MAPK1,PRKCD,PRKCH,PIK3R2,RAC3 Tumor AKT2,PLD3,MAPK1,RPS6KB2,TLN1,VASP,PTEN Prostate Cancer FAP PIK3C2B,SOS1,MAP2K1 Prostate Cancer FGFR2 Prostate Cancer Hyper FOXO1,MAPK1,CREBBP,PIK3R2,NFKB2 Prostate Cancer Tumor AKT2,MTOR,NFKBIA,TFDP1,MAPK1,PIK3C2A,PIK3 C3,SOS2,CTNNB1,PTEN Phosphatidylglycerol Biosynthesis II (Non-plastidic) Phosphatidylglycerol Biosynthesis II (Non-plastidic) Phosphatidylglycerol Biosynthesis II (Non-plastidic) Virus Entry via Endocytic Pathways Virus Entry via Endocytic Pathways FAP TAZ LPCAT4 Hyper TAMM41,LPCAT2,AGPAT3 FAP PIK3C2B,ITGA1,PRKD FGFR2,AP1B1
200 Virus Entry via Endocytic Pathways Virus Entry via Endocytic Pathways Hyper DNM1,FLNB,PRKCD,PRKCH,PIK3R2,RAC3 Tumor FLNB,AP1G2,PIK3C2A,HLA- A,PIK3C3,AP2B1,ITGA1,DNM2 Remodeling of Epithelial Adherens Junctions Remodeling of Epithelial Adherens Junctions Remodeling of Epithelial Adherens Junctions Remodeling of Epithelial Adherens Junctions 4-1BB in T Lymphocytes 4-1BB in T Lymphocytes 4-1BB in T Lymphocytes Inhibition of Angiogenesis by TSP1 Inhibition of Angiogenesis by TSP1 Inhibition of Angiogenesis by TSP1 Role of JAK2 in Hormone-like Cytokine Role of JAK2 in Hormone-like Cytokine Role of JAK2 in Hormone-like Cytokine Role of JAK2 in Hormone-like Cytokine FAP ACTR2,CLIP1 Hyper MAPRE DNM1,TUBB3,TUBB6,TUBG1,HGS,ACTN1,CLIP1 Tumor TUBB3,TUBB4B,ZYX,CTNNB1,DNM2,CTNND1 FAP MAP2K1 Hyper MAPK1,NFKB2 Tumor NFKBIA,JUN,MAPK1 FAP CD47 Hyper HSPG2,TGFBR1,MAPK1 Tumor AKT2,JUN,MAPK1 FAP STAT TYK2,STAT3 Hyper STAT5A,HLTF Tumor PTPN1
201 Aryl Hydrocarbon Receptor Hyper NCOA7,MAPK1,DHFR,NFKB2,NCOR2,RXRB,SMA RCA4 Aryl Hydrocarbon Receptor Tumor TRIP11,RBL2,JUN,TFDP1,MAPK1,APAF1,CDK4,AL DH18A1,SMARCA4,MCM7 LXR/RXR Activation Hyper SCD,TNFRSF1A,SREBF1,FASN,ABCG1,IRF3,NFK B2,NCOR2,RXRB LXR/RXR Activation Tumor SREBF1,FASN,NCOR1,HMGCR VDR/RXR Activation VDR/RXR Activation Hyper KLF FOXO1,PRKCD,NCOA1,PRKCH,CEBPB,NCOR2,H R,RXRB,KLF4,PSMC5 Caveolar-mediated Endocytosis IL-8 Tumor FLNB,HLA-A,PTPN1,ITGA1,DNM2 Hyper PTK2,ROCK1,RHOQ,MAPK1,PRKCD,PRKCH,PIK3 R2,RAC3 IL-8 Tumor ROCK1,BRAF,AKT2,ARRB2,MTOR,PLD3,JUN,MAP K1,PIK3C2A,PIK3C3,RACK1,VASP Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses Hyper OAS2,FGFR2,TLR MAPK1,TICAM1,PRKCD,PRKCH,IRF3,PIK3R2,NFK B2,OAS3 Tumor MAPK1,PIK3C2A,PIK3C3,C1QA
202 TREM TLR3,STAT3 TREM1 Hyper SIGIRR,STAT5A,NLRC5,MAPK1,NFKB2 TREM1 Tumor AKT2,MAPK1,CASP5 Lymphotoxin β Receptor Lymphotoxin β Receptor Lymphotoxin β Receptor FGFR2 Hyper MAPK1,CREBBP,PIK3R2,NFKB2,BIRC2 Tumor AKT2,NFKBIA,MAPK1,PIK3C2A,PIK3C3,APAF1 IL FGFR2 IL-17 Hyper MAPK1,CEBPB,PIK3R2 IL-17 Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,MUC5B CTLA4 in Cytotoxic T Lymphocytes CTLA4 in Cytotoxic T Lymphocytes Reelin in Neurons Reelin in Neurons FGFR2,AP1B1 Tumor AKT2,AP1G2,PIK3C2A,HLA-A,PIK3C3,AP2B1 Hyper FGFR2,ARHGEF1,ARHGEF ARHGEF5,MAP3K11,PIK3R2,MAPK8IP3,FRK Reelin in Neurons Tumor PIK3C2A,PIK3C3,ARHGEF1,ITGA1 Relaxin Tumor BRAF,AKT2,NFKBIA,JUN,MAPK1,PIK3C2A,PIK3C3, RACK1,NPR2
203 Agrin Interactions at Neuromuscular Junction Agrin Interactions at Neuromuscular Junction Docosahexaenoic Acid (DHA) Docosahexaenoic Acid (DHA) Docosahexaenoic Acid (DHA) Antiproliferative Role of Somatostatin Receptor 2 Antiproliferative Role of Somatostatin Receptor 2 Hyper PTK2,MAPK1,DVL1,ERBB2,AGRN,RAC3 Tumor JUN,MAPK1,ARHGEF7,DVL1,ITGA FGFR2 Hyper FOXO1,PIK3R2,PNPLA2 Tumor AKT2,PIK3C2A,PIK3C3,APAF FGFR2 Tumor BRAF,MAPK1,PIK3C2A,PIK3C3,RACK1,NPR2 Germ Cell-Sertoli Cell Junction Germ Cell-Sertoli Cell Junction Hyper KEAP1,FGFR TUBB3,TGFBR1,MAP3K11,MAPK1,TNFRSF1A,TUB G1,NECTIN3,RAC3,PTK2,RHOQ,TUBB6,SORBS1, KEAP1,PIK3R2,ACTN1,NECTIN2 Germ Cell-Sertoli Cell Junction Tumor TUBB3,MAPK1,PIK3C2A,TUBB4B,PIK3C3,NECTIN 3,ILK,ZYX,CLINT1,CTNNB1,CTNND1 Small Cell Lung Cancer Small Cell Lung Cancer Hyper FGFR2,SKP PTK2,PIK3R2,NFKB2,RXRB,BIRC2
204 Small Cell Lung Cancer Tumor AKT2,NFKBIA,TFDP1,PIK3C2A,PIK3C3,APAF1,CD K4,SKP2,PTEN Type II Diabetes Mellitus Type II Diabetes Mellitus Hyper FGFR2,SMPD PRKAB2,MAPK1,TNFRSF1A,PRKCD,PRKCH,CEBP B,PIK3R2,NFKB2,NSMAF Type II Diabetes Mellitus Tumor AKT2,ACSL3,MTOR,NFKBIA,MAPK1,PIK3C2A,PIK3 C3,ADIPOR1 Myc Mediated Apoptosis Myc Mediated Apoptosis Pancreatic Adenocarcinoma Pancreatic Adenocarcinoma Pancreatic Adenocarcinoma FGFR2 Tumor AKT2,PIK3C2A,PIK3C3,SOS2,APAF TYK2,FGFR2,STAT3,PLD1 Hyper E2F4,TGFBR1,MAPK1,ERBB2,PIK3R2,NFKB2 Tumor SMAD2,AKT2,PLD3,TFDP1,MAPK1,PIK3C2A,PIK3 C3,CDK4,NOTCH1 Sphingosine-1-phosphate Sphingosine-1-phosphate Hyper PLCD3,FGFR2,SMPD PTK2,PLCD3,RHOQ,MAPK1,CASP2,ADCY3,ADCY 6,PLCB3,PIK3R2 Sphingosine-1-phosphate Tumor AKT2,MAPK1,PIK3C2A,PIK3C3,PLCH1,CASP5
205 Systemic Lupus Erythematosus Tumor AKT2,MTOR,JUN,MAPK1,PIK3C2A,HLA- A,PIK3C3,PRPF8,SOS2,SNRNP200,SNRNP40 ILK Hyper PTK2,NCK2,FLNB,MYH9,RHOQ,MAPK1,TNFRSF1 A,CREBBP,PIK3R2,NFKB2,ACTN1 ILK Tumor PARVB,FLNB,AKT2,MTOR,JUN,MAPK1,PIK3C2A,P IK3C3,ILK,CTNNB1,PTEN AMPK AMPK Hyper PBRM1,CPT1A,CPT1B,FGFR TSC1,PRKAB2,MAPK1,CREBBP,MT8,SMARCA4,FOXO1,FASN,SMARCB1,TSC2,SIRT1,PIK3R2,HLT F AMPK Tumor PBRM1,AKT2,MTOR,MAPK1,PIK3C2A,EEF2,PIK3C 3,FASN,ARID2,HMGCR,SMARCA4,PFKM Ovarian Cancer Tumor AKT2,TFDP1,PIK3C2A,MAPK1,DVL1,CDK4,MLH1,P TEN,BRAF,MTOR,PIK3C3,RPS6KB2,MSH6,CTNNB 1 Estrogen-Dependent Breast Cancer Estrogen-Dependent Breast Cancer Hyper FGFR STAT5A,MAPK1,CREBBP,PIK3R2,NFKB2
206 Estrogen-Dependent Breast Cancer Role of PI3K/AKT in the Pathogenesis of Influenza Role of PI3K/AKT in the Pathogenesis of Influenza Role of PI3K/AKT in the Pathogenesis of Influenza Role of Tissue Factor in Cancer Sertoli Cell-Sertoli Cell Junction Sertoli Cell-Sertoli Cell Junction Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,HSD17B FGFR2 Hyper MAPK1,IRF3,PIK3R2,NFKB2 Tumor AKT2,NFKBIA,MAPK1,PIK3C2A,PIK3C3,MLH1 Tumor AKT2,ARRB2,MTOR,MAPK1,PIK3C2A,PIK3C3,PTE N DLG1,KEAP1 Hyper EPB41,TUBB3,MAP3K11,MAPK1,TUBB6,SORBS1, TNFRSF1A,KEAP1,NECTIN3,TUBG1,ACTN1,NECT IN2 Sertoli Cell-Sertoli Cell Junction Tumor AKT2,TUBB3,JUN,MAPK1,TUBB4B,NECTIN3,ILK,S YMPK,CLINT1,CTNNB1,PTEN inos TYK2 inos Hyper MAPK1,CREBBP,NFKB2 inos Tumor NFKBIA,JUN,MAPK1,IRF1 Cell Cycle: G1/S Checkpoint Regulation Cell Cycle: G1/S Checkpoint Regulation Hyper SKP E2F4,FOXO1,HDAC8,CDKN2C,HDAC5
207 Cell Cycle: G1/S Checkpoint Regulation Nucleotide Excision Repair Pathway Nucleotide Excision Repair Pathway PI3K/AKT Tumor RBL2,TFDP1,HDAC1,CDK4,SKP2 Hyper XPC,XPA Tumor POLR2C,POLR2A,ERCC3 Hyper TSC1,FOXO1,MAPK1,TSC2,PIK3R2,NFKB2,THEM4 PI3K/AKT Tumor AKT2,MTOR,NFKBIA,MAPK1,SOS2,ILK,RPS6KB2, CTNNB1,PTEN IL TYK2,FGFR2 IL-4 Tumor STAT6,AKT2,MTOR,PIK3C2A,HLA- A,PIK3C3,SOS2,RPS6KB2 Phagosome Maturation Phagosome Maturation Hyper ATP6V0A2,ATP6V0D CALR,TUBB3,TUBB6,TUBE1,CTSO,VPS33A,TUBG 1,NAPG,VPS37C,CTSF Phagosome Maturation Tumor DYNC1H1,CTSA,TUBB3,TUBB4B,HLA- A,PIK3C3,ATP6V0D1,PIKFYVE,ATP6V0A1 Cancer Drug Resistance By Drug Efflux Cancer Drug Resistance By Drug Efflux Cancer Drug Resistance By Drug Efflux ABCG2 Hyper FOXO1,MAPK1,PIK3R2 Tumor BRAF,AKT2,MAPK1,PTEN
208 IL-7 Pathway FGFR2 IL-7 Pathway Hyper PTK2,STAT5A,FOXO1,MAPK1,PIK3R2 IL-7 Pathway Tumor AKT2,JUN,MAPK1,PIK3C2A,PIK3C3,SOS2 Opioid Pathway Hyper CAMK1,MAPK1,ITPR2,PRKCD,CREBBP,ADCY3,G RK6,ADCY6,PRKCH,NFKB2,FRK,RAC3 Opioid Pathway Tumor BRAF,AKT2,ARRB2,NFKBIA,AP1G2,MAPK1,AP2B1,ITPR3,SOS2,RPS6KB2,CTNNB1 IL-15 Production TYK2,MST1R IL-15 Production Hyper PTK2,MAP3K11,IRF3,NFKB2,FRK IL-15 Production Tumor IRF1 Antiproliferative Role of TOB in T Cell Antiproliferative Role of TOB in T Cell Antiproliferative Role of TOB in T Cell IL-17A in Fibroblasts IL-17A in Fibroblasts Estrogen-mediated S-phase Entry SKP2 Hyper TGFBR1,MAPK1,PABPC4 Tumor SMAD2,MAPK1,SKP2 Hyper MAPK1,CEBPB,NFKB2 Tumor NFKBIA,JUN,MAPK1 Hyper SKP2 Estrogen-mediated S-phase E2F4 Entry Estrogen-mediated S-phase Tumor TFDP1,CDK4,SKP2
209 Entry Thio-molybdenum Cofactor Tumor MOCOS Biosynthesis 2-ketoglutarate Tumor DT,OGDH Dehydrogenase Complex Palmitate Biosynthesis I Hyper FASN (Animals) Palmitate Biosynthesis I Tumor FASN (Animals) Anandamide Degradation Tumor FAAH2 Fatty Acid Biosynthesis Hyper FASN Initiation II Fatty Acid Biosynthesis Tumor FASN Initiation II Zymosterol Biosynthesis Hyper LBR Zymosterol Biosynthesis Tumor HSD17B7,LBR BER pathway Hyper LIG1 BER pathway Tumor POLE,LIG3 Ingenuity Pathway Analysis of predicted neoantigens with binding affinity <500nM, allele frequency >5% and mrna expression >10 TPM. Pathways with P-value<0.05 in either FAP, s or Tumors are displayed.
210 etable 11. Germline and somatic mutation detected in FAP and polyps Sample ID Syndrome Patient Germline Mutation Gene cdna Protein ID Validation FAP_G2 FAP FAP_1 Germline APC c.1880dupa p.ala630* Yes Germline APC c.1880dupa p.ala630* Yes FAP_G3 FAP FAP_1 Somatic p.(phe1491_ala1492delinsleu APC c.4473_4474delinsgc Pro) Somatic KRAS c.519t>c p.(=) Germline APC c.3810t>a p.cys1270* Yes FAP_G53 FAP FAP_2 Somatic APC c.2438a>g p.(asn813ser) Somatic APC c.4348c>t p.(arg1450*) FAP_G55 FAP FAP_2 Germline APC c.3810t>a p.cys1270* Yes FAP_G5 FAP FAP_3 Germline APC del exon 15 p.? No Somatic APC LOH 5q FAP_G6 FAP FAP_3 Germline APC del exon 16 p.? No Germline APC c.622c>t p.gln208* Yes FAP_G13 FAP FAP_4 Somatic APC c.4057g>t p.(glu1353*) Somatic APC c.4135g>t p.(glu1379*) FAP_G14 FAP FAP_4 Germline APC c.622c>t p.gln208* No Germline APC c.622c>t p.gln208* Yes FAP_G15 FAP FAP_4 Somatic APC c.4189_4190del p.(arg1399phefs*9) Somatic APC LOH 5q FAP_G25 FAP FAP_5 Germline APC del exons 8-10 p.? No FAP_G29 FAP FAP_6 Germline APC c.1658g>a p.trp553* Yes FAP_G39 FAP FAP_7 Germline APC c.477c>g p.tyr159* No FAP_G47 FAP FAP_8 Germline APC c.4733_4734del p.cys1578tyrfs*13 No Somatic APC LOH 5q FAP_G48 FAP FAP_8 Germline APC c.4733_4734del p.cys1578tyrfs*14 No Germline APC c.847c>t p.arg283* No FAP_G67 FAP FAP_9 Somatic APC c.4135g>t p.(glu1379*) Somatic KRAS c.35c>a p.(gly12val) FAP_G61 FAP FAP_10 Germline APC c.3810t>a p.cys1270* Yes _EBL1 _1 Germline MSH6 c.3744_3773del p.his1248_ser1257del Yes _EBL3 _2 Germline MSH2 c.687del p.ala230leufs*16 Yes _EBL5 _3 Germline PMS2 del exon 14 p.? No
211 Somatic MLH1 c.342_343del p.(ile115tyrfs*6) _EBL7 _4 Germline MSH2 c.1034g>a p.trp345* Yes Germline MSH2 c g>a p.? No _EBL9 _5 Somatic MSH6 c.1340t>c p.(leu447pro) Somatic MSH6 c.3410t>c p.(met1137thr) Germline MSH2 del exons 1-6 p.? No Somatic MSH2 c.2172g>a p.(=) _EBL11 _6 Somatic APC c.2626c>t Somatic KRAS c.485a>g p.(glu162gly) Somatic CIITA c.1436c>t p.a479v _EBL13 _7 Germline MSH2 c.1216c>t p.arg406* Yes _EBL18 _8 Germline MSH6 c.3238_3239del p.leu1080valfs*12 No Germline MSH6 c.3860_3861insatta p.y1287* Yes _EBL26 _9 Somatic MSH2 c.2646_2647del p.(lys882asnfs*16) Somatic APC c.4348c>t p.(arg1450*) Germline MSH6 c.2645_2653del p.phe882* Yes _EBL20 _10 Somatic APC c.4234g>t p. Somatic APC LOH 5q _EBL23 _11 Germline MSH6 c.3699_3702del p.lys1233asnfs*6 Yes Germline MLH1 c.1279c>t p.gln427* Yes Somatic APC c.c4549t p.q1517x Somatic BRAF c.1208delc p.p403fs _T3 _12 Somatic CTNNB1 c.c134t p.s45f Somatic AXIN1 c.1922dela p.k641fs Somatic RFX5 c.56delc p.p19fs Somatic RFXAP c.297delg p.p99fs Germline MLH1 c.1918c>t p.pro640ser Yes Somatic APC c.2540dupa p.e847fs Somatic KRAS c.g38a p.g13d _T2 _13 Somatic MLH1 c.c350t p.t117m Somatic c.666_667insgatggag MSH6 p.d222fs G Somatic c.668_669insggcacaa MSH6 CTTACGTAAC p.n223_e224delinskaqltx _T1 _14 Germline MLH1 c.2059c>t p.arg687trp Yes Mutational data derived from RNA-seq analysis. Sample FAP_G53 and _EBL26 harbored APC LOH in 5q in addition to the somatic mutations.
212 efigure 1 A Tumor T-cell Signature Enrichment efigure 1. A. Immune profile of Lynch syndrome premalignancy and carcinomas. RNA expression levels of immune-related genes invovled in CD4, Th1/Tc1, CTL, checkpoint response, TH17, Treg, Proinflammation and Metabolism comparing polyps and tumors; B. T-cell signature enrichment score comparing andfap polyps, tumors.the graphs display mean for each group and statistically significant difference between each group using ANOVA and Tukey s Test, and multiple comparisons by Benjamini & Hochberg method. (*P<0.05, **P<0.01, ***P<0.001).
213 efigure 2 RNA mutation rate (Mutations/Mb) DNA mutation rate (Mutations/Mb) R 2 = P-value < efigure 2. Correlation of DNA and RNA mutation rate in samples from TCGA. A total of 47 Stage I and II colorectal tumors with known MSI and hypermutation status from TCGA were selected for analysis.
214 efigure 3
215 efigure 4 CD4 FOXP3 efigure 4. Immunohistochemical staining of CD4 and FOXP3. Representative hypermutant Lynch syndrome polyp showing abundant infiltration by CD4 positive/foxp3 positive T-cells.
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Single-site association results for 136 SCARB1 genotyped variants with HDL-C.
 Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Supplemental file 3. All 306 mapped IDs collected by IPA program. Supplemental file 6. The functions and main focused genes in each network.
 LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
J. Dairy Sci. 93: doi: /jds American Dairy Science Association, 2010.
 Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Prostate-targeted radiosensitization via aptamer-shrna chimeras in human. tumor xenografts
 SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
PRELIMINARY RESULTS FROM THE FIRST TRIALS OF THE GENSINGEN BRACE (BY Dr. WEISS) IN GREECE
 PRELIMINARY RESULTS FROM THE FIRST TRIALS OF THE GENSINGEN BRACE (BY Dr. WEISS) IN GREECE Objective: The Gensingen brace is a new asymmetrical CAD/CAM Cheneau type brace, with very promising published
PRELIMINARY RESULTS FROM THE FIRST TRIALS OF THE GENSINGEN BRACE (BY Dr. WEISS) IN GREECE Objective: The Gensingen brace is a new asymmetrical CAD/CAM Cheneau type brace, with very promising published
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
Μοριακή Διαγνωστική. Σύγχρονες Εφαρμογές Μοριακής Διαγνωστικής. Κλινική Χημεία ΣΤ Εξάμηνο - Παραδόσεις
 Εθνικό και Καποδιστριακό Πανεπιστήμιο Αθηνών Τμήματα Βιολογίας Σ.Θ.Ε. Τομέας Βιοχημείας και Μοριακής Βιολογίας Μοριακή Διαγνωστική Σύγχρονες Εφαρμογές Μοριακής Διαγνωστικής Κλινική Χημεία ΣΤ Εξάμηνο -
Εθνικό και Καποδιστριακό Πανεπιστήμιο Αθηνών Τμήματα Βιολογίας Σ.Θ.Ε. Τομέας Βιοχημείας και Μοριακής Βιολογίας Μοριακή Διαγνωστική Σύγχρονες Εφαρμογές Μοριακής Διαγνωστικής Κλινική Χημεία ΣΤ Εξάμηνο -
Nature Medicine doi: /nm.2772
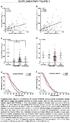 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
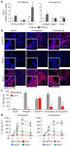 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
Figure 3 Three observations (Vp, Vs and density isosurfaces) intersecting in the PLF space. Solutions exist at the two indicated points.
 φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
φ φ φ φ Figure 1 Resampling of a rock-physics model from velocity-porosity to lithology-porosity space. C i are model results for various clay contents. φ ρ ρ δ Figure 2 Bulk modulus constraint cube in
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Προενταξιακός ασθενής - Επιλογή μεθόδου κάθαρσης
 Προενταξιακός ασθενής - Επιλογή μεθόδου κάθαρσης Παπαχρήστου Ευάγγελος Επίκουρος Καθηγητής Παθολογίας Νεφρολογίας Ιατρική Σχολή Πανεπιστημίου Πατρών Χρόνια Νεφρική Νόσος (ΧΝΝ) Διαταραχή της δομής ή της
Προενταξιακός ασθενής - Επιλογή μεθόδου κάθαρσης Παπαχρήστου Ευάγγελος Επίκουρος Καθηγητής Παθολογίας Νεφρολογίας Ιατρική Σχολή Πανεπιστημίου Πατρών Χρόνια Νεφρική Νόσος (ΧΝΝ) Διαταραχή της δομής ή της
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
athanasiadis@rhodes.aegean.gr , -.
 παιδαγωγικά ρεύµατα στο Αιγαίο Προσκήνιο 88 - * athanasiadis@rhodes.aegean.gr -., -.. Abstract The aim of this survey is to show how students of the three last school classes of the Primary School evaluated
παιδαγωγικά ρεύµατα στο Αιγαίο Προσκήνιο 88 - * athanasiadis@rhodes.aegean.gr -., -.. Abstract The aim of this survey is to show how students of the three last school classes of the Primary School evaluated
 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
Aquinas College. Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET
 Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Statistical Inference I Locally most powerful tests
 Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Καρδιακή Συχνότητα και Πρόσληψη Οξυγόνου Ατόμων Μέσης Ηλικίας κατά την Εκτέλεση Ελληνικών Παραδοσιακών Χορών
 Ερευνητική Αναζητήσεις στη Φυσική Αγωγή & τον Αθλητισμό τόμος 6 (3), 329 339 Δημοσιεύτηκε: 31 Δεκεμβρίου 2008 Inquiries in Sport & Physical Education Volume 6 (3), 329-339 Released: December 31, 2008 www.hape.gr/emag.asp
Ερευνητική Αναζητήσεις στη Φυσική Αγωγή & τον Αθλητισμό τόμος 6 (3), 329 339 Δημοσιεύτηκε: 31 Δεκεμβρίου 2008 Inquiries in Sport & Physical Education Volume 6 (3), 329-339 Released: December 31, 2008 www.hape.gr/emag.asp
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών
 Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
derivation of the Laplacian from rectangular to spherical coordinates
 derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine
 Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
Μαντζούνη, Πιπερίγκου, Χατζή. ΒΙΟΣΤΑΤΙΣΤΙΚΗ Εργαστήριο 5 ο
 Κατανομές Στατιστικών Συναρτήσεων Δύο δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,...,Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ ) S σ Τ ( Χ,Y)
Κατανομές Στατιστικών Συναρτήσεων Δύο δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,...,Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ ) S σ Τ ( Χ,Y)
Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks. Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks
 Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
EPL 603 TOPICS IN SOFTWARE ENGINEERING. Lab 5: Component Adaptation Environment (COPE)
 EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Περίληψη (Executive Summary)
 1 Περίληψη (Executive Summary) Η παρούσα διπλωματική εργασία έχει ως αντικείμενο την "Αγοραστική/ καταναλωτική συμπεριφορά. Η περίπτωση των Σπετσών" Κύριος σκοπός της διπλωματικής εργασίας είναι η διερεύνηση
1 Περίληψη (Executive Summary) Η παρούσα διπλωματική εργασία έχει ως αντικείμενο την "Αγοραστική/ καταναλωτική συμπεριφορά. Η περίπτωση των Σπετσών" Κύριος σκοπός της διπλωματικής εργασίας είναι η διερεύνηση
Sampling Basics (1B) Young Won Lim 9/21/13
 Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
Sampling Basics (1B) Copyright (c) 2009-2013 Young W. Lim. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any
ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE ΑΠΟ ΑΤΟΜΑ ΜΕ ΤΥΦΛΩΣΗ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
Supplemental Table 1. ICD-9-CM codes and ATC codes used in this study
 Supplemental Table 1. ICD-9-CM codes and ATC codes used in this study Comorbidities ICD-9-CM codes Medication ATC codes Diabetes 250 Angiotensin-converting enzyme inhibitors C09AA Ischemic heart disease
Supplemental Table 1. ICD-9-CM codes and ATC codes used in this study Comorbidities ICD-9-CM codes Medication ATC codes Diabetes 250 Angiotensin-converting enzyme inhibitors C09AA Ischemic heart disease
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΜΔΗΕΟΝ ΤΜΠΛΔΓΜΑ ΗΣΟΤΜΒΑΣΟΣΖΣΑ
 ΜΔΗΕΟΝ ΤΜΠΛΔΓΜΑ ΗΣΟΤΜΒΑΣΟΣΖΣΑ 5 Σε όια ζρεδόλ ηα ζπνλδπισηά, ηα νπνία έρνπλ έσο ζήκεξα κειεηεζεί θαη ηδίσο ζηα ζειαζηηθά, ζπκπεξηιακβαλνκέλνπ θαη ηνπ αλζξώπνπ, έρεη δηαπηζησζεί ε ύπαξμε κηαο ζηελά ζπλδεδεκέλεο
ΜΔΗΕΟΝ ΤΜΠΛΔΓΜΑ ΗΣΟΤΜΒΑΣΟΣΖΣΑ 5 Σε όια ζρεδόλ ηα ζπνλδπισηά, ηα νπνία έρνπλ έσο ζήκεξα κειεηεζεί θαη ηδίσο ζηα ζειαζηηθά, ζπκπεξηιακβαλνκέλνπ θαη ηνπ αλζξώπνπ, έρεη δηαπηζησζεί ε ύπαξμε κηαο ζηελά ζπλδεδεκέλεο
Ανάκτηση Πληροφορίας
 Ανάκτηση Πληροφορίας Αποτίμηση Αποτελεσματικότητας Μέτρα Απόδοσης Precision = # σχετικών κειμένων που επιστρέφονται # κειμένων που επιστρέφονται Recall = # σχετικών κειμένων που επιστρέφονται # συνολικών
Ανάκτηση Πληροφορίας Αποτίμηση Αποτελεσματικότητας Μέτρα Απόδοσης Precision = # σχετικών κειμένων που επιστρέφονται # κειμένων που επιστρέφονται Recall = # σχετικών κειμένων που επιστρέφονται # συνολικών
Table 2 Suggested prediction methods to use for a given set of available input parameters per each examined soil hydraulic property) a.
 Table 2 Suggested prediction methods to use for a given set of available input parameters per each examined soil hydraulic property) a. Reference number of model indicated in Table 1 Predicted soil hydraulic
Table 2 Suggested prediction methods to use for a given set of available input parameters per each examined soil hydraulic property) a. Reference number of model indicated in Table 1 Predicted soil hydraulic
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ Πτυχιακή εργασία ΓΝΩΣΕΙΣ ΚΑΙ ΣΤΑΣΕΙΣ ΝΟΣΗΛΕΥΤΩΝ ΠΡΟΣ ΤΟΥΣ ΦΟΡΕΙΣ ΜΕ ΣΥΝΔΡΟΜΟ ΕΠΙΚΤΗΤΗΣ ΑΝΟΣΟΑΝΕΠΑΡΚΕΙΑΣ (AIDS) Αλέξης Δημήτρη Α.Φ.Τ: 20085675385 Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΤΜΗΜΑ ΝΟΣΗΛΕΥΤΙΚΗΣ Πτυχιακή εργασία ΓΝΩΣΕΙΣ ΚΑΙ ΣΤΑΣΕΙΣ ΝΟΣΗΛΕΥΤΩΝ ΠΡΟΣ ΤΟΥΣ ΦΟΡΕΙΣ ΜΕ ΣΥΝΔΡΟΜΟ ΕΠΙΚΤΗΤΗΣ ΑΝΟΣΟΑΝΕΠΑΡΚΕΙΑΣ (AIDS) Αλέξης Δημήτρη Α.Φ.Τ: 20085675385 Λεμεσός
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Consolidated Drained
 Consolidated Drained q, 8 6 Max. Shear c' =.185 φ' =.8 tan φ' =.69 Deviator, 8 6 6 8 1 1 p', 5 1 15 5 Axial, Symbol Sample ID Depth Test Number Height, in Diameter, in Moisture Content (from Cuttings),
Consolidated Drained q, 8 6 Max. Shear c' =.185 φ' =.8 tan φ' =.69 Deviator, 8 6 6 8 1 1 p', 5 1 15 5 Axial, Symbol Sample ID Depth Test Number Height, in Diameter, in Moisture Content (from Cuttings),
ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ & ΠΕΡΙΒΑΛΛΟΝΤΟΣ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΠΑΡΑΡΤΗΜΑ ΧΑΝΙΩΝ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ & ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΤΟΜΕΑΣ ΠΕΡΙΒΑΛΛΟΝΤΙΚΗΣ ΤΕΧΝΟΛΟΓΙΑΣ ΕΡΓΑΣΤΗΡΙΟ ΠΕΡΙΒΑΛΛΟΝΤΙΚΗΣ ΧΗΜΕΙΑΣ & ΒΙΟΧΗΜΙΚΩΝ ΙΕΡΓΑΣΙΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΠΑΡΑΡΤΗΜΑ ΧΑΝΙΩΝ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ & ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΤΟΜΕΑΣ ΠΕΡΙΒΑΛΛΟΝΤΙΚΗΣ ΤΕΧΝΟΛΟΓΙΑΣ ΕΡΓΑΣΤΗΡΙΟ ΠΕΡΙΒΑΛΛΟΝΤΙΚΗΣ ΧΗΜΕΙΑΣ & ΒΙΟΧΗΜΙΚΩΝ ΙΕΡΓΑΣΙΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ
Μενύχτα, Πιπερίγκου, Σαββάτης. ΒΙΟΣΤΑΤΙΣΤΙΚΗ Εργαστήριο 5 ο
 Κατανομές Στατιστικών Συναρτήσεων Δύο ανεξάρτητα δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,..., Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ )
Κατανομές Στατιστικών Συναρτήσεων Δύο ανεξάρτητα δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,..., Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ )
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ. Τα γνωστικά επίπεδα των επαγγελματιών υγείας Στην ανοσοποίηση κατά του ιού της γρίπης Σε δομές του νομού Λάρισας
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΡΩΤΟΒΑΘΜΙΑ ΦΡΟΝΤΙΔΑ ΥΓΕΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Τα γνωστικά επίπεδα των επαγγελματιών υγείας Στην ανοσοποίηση
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΡΩΤΟΒΑΘΜΙΑ ΦΡΟΝΤΙΔΑ ΥΓΕΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Τα γνωστικά επίπεδα των επαγγελματιών υγείας Στην ανοσοποίηση
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
TABLE OF CONTENTS Page
 TABLE OF CONTENTS Page Acknowledgements... VI Declaration... VII List of abbreviations... VIII 1. INTRODUCTION... 1 2. BASIC PRINCIPLES... 3 2.1. Hereditary Colorectal Cancer... 3 2.1.1. Differential diagnosis...
TABLE OF CONTENTS Page Acknowledgements... VI Declaration... VII List of abbreviations... VIII 1. INTRODUCTION... 1 2. BASIC PRINCIPLES... 3 2.1. Hereditary Colorectal Cancer... 3 2.1.1. Differential diagnosis...
Chapter 1. Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes
 Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Σχέση µεταξύ της Μεθόδου των ερµατοπτυχών και της Βιοηλεκτρικής Αντίστασης στον Υπολογισµό του Ποσοστού Σωµατικού Λίπους
 Ερευνητική Aναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµότόµος 1(3), 244 251 ηµοσιεύτηκε: 2 8 εκεµβρίου 2003 Inquiries in Sport & Physical Education Volume 1(3), 244 251 Released: December 28, 2003 www.hape.gr/emag.asp
Ερευνητική Aναζητήσεις στη Φυσική Αγωγή & τον Αθλητισµότόµος 1(3), 244 251 ηµοσιεύτηκε: 2 8 εκεµβρίου 2003 Inquiries in Sport & Physical Education Volume 1(3), 244 251 Released: December 28, 2003 www.hape.gr/emag.asp
Statistics 104: Quantitative Methods for Economics Formula and Theorem Review
 Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
NCBI ID Symbol 2 A2M A4GALT A4GNT 8086 AAAS 13 AADAC AADACL AAGAB 15 AANAT 16 AARS AARS AASS 18 ABAT 19 ABCA1
 NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων. Εξάμηνο 7 ο
 Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων Εξάμηνο 7 ο Procedures and Functions Stored procedures and functions are named blocks of code that enable you to group and organize a series of SQL and PL/SQL
Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων Εξάμηνο 7 ο Procedures and Functions Stored procedures and functions are named blocks of code that enable you to group and organize a series of SQL and PL/SQL
Τ.Ε.Ι. ΔΥΤΙΚΗΣ ΜΑΚΕΔΟΝΙΑΣ ΠΑΡΑΡΤΗΜΑ ΚΑΣΤΟΡΙΑΣ ΤΜΗΜΑ ΔΗΜΟΣΙΩΝ ΣΧΕΣΕΩΝ & ΕΠΙΚΟΙΝΩΝΙΑΣ
 Τ.Ε.Ι. ΔΥΤΙΚΗΣ ΜΑΚΕΔΟΝΙΑΣ ΠΑΡΑΡΤΗΜΑ ΚΑΣΤΟΡΙΑΣ ΤΜΗΜΑ ΔΗΜΟΣΙΩΝ ΣΧΕΣΕΩΝ & ΕΠΙΚΟΙΝΩΝΙΑΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η προβολή επιστημονικών θεμάτων από τα ελληνικά ΜΜΕ : Η κάλυψή τους στον ελληνικό ημερήσιο τύπο Σαραλιώτου
Τ.Ε.Ι. ΔΥΤΙΚΗΣ ΜΑΚΕΔΟΝΙΑΣ ΠΑΡΑΡΤΗΜΑ ΚΑΣΤΟΡΙΑΣ ΤΜΗΜΑ ΔΗΜΟΣΙΩΝ ΣΧΕΣΕΩΝ & ΕΠΙΚΟΙΝΩΝΙΑΣ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η προβολή επιστημονικών θεμάτων από τα ελληνικά ΜΜΕ : Η κάλυψή τους στον ελληνικό ημερήσιο τύπο Σαραλιώτου
Παιδιατρική ΒΟΡΕΙΟΥ ΕΛΛΑΔΟΣ, 23, 3. Γ Παιδιατρική Κλινική, Αριστοτέλειο Πανεπιστήμιο, Ιπποκράτειο Νοσοκομείο Θεσσαλονίκης, 2
 162 Σύγκριση της επίδρασης της Μοντελουκάστης και των Εισπνεομένων Στεροειδών στο Εκπνεόμενο Μονοξείδιο του Αζώτου (eno) και την αναπνευστική λειτουργία σε ασθματικά παιδιά Ε. Χατζηαγόρου 1, Β. Αβραμίδου
162 Σύγκριση της επίδρασης της Μοντελουκάστης και των Εισπνεομένων Στεροειδών στο Εκπνεόμενο Μονοξείδιο του Αζώτου (eno) και την αναπνευστική λειτουργία σε ασθματικά παιδιά Ε. Χατζηαγόρου 1, Β. Αβραμίδου
Solution Series 9. i=1 x i and i=1 x i.
 Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
Supplementary Materials: A Preliminary Link between Hydroxylated Metabolites of Polychlorinated Biphenyls and Free Thyroxin in Humans
 S1 of S11 Supplementary Materials: A Preliminary Link between Hydroxylated Metabolites of Polychlorinated Biphenyls and Free Thyroxin in Humans Eveline Dirinck, Alin C. Dirtu, Govindan Malarvannan, Adrian
S1 of S11 Supplementary Materials: A Preliminary Link between Hydroxylated Metabolites of Polychlorinated Biphenyls and Free Thyroxin in Humans Eveline Dirinck, Alin C. Dirtu, Govindan Malarvannan, Adrian
Repeated measures Επαναληπτικές μετρήσεις
 ΠΡΟΒΛΗΜΑ Στο αρχείο δεδομένων diavitis.sav καταγράφεται η ποσότητα γλυκόζης στο αίμα 10 ασθενών στην αρχή της χορήγησης μιας θεραπείας, μετά από ένα μήνα και μετά από δύο μήνες. Μελετήστε την επίδραση
ΠΡΟΒΛΗΜΑ Στο αρχείο δεδομένων diavitis.sav καταγράφεται η ποσότητα γλυκόζης στο αίμα 10 ασθενών στην αρχή της χορήγησης μιας θεραπείας, μετά από ένα μήνα και μετά από δύο μήνες. Μελετήστε την επίδραση
Relationship between Connective Tissue Diseases and Thyroid Diseases
 CHINESE JOURNAL OF ALLERGY & CLINICAL IMMUNOLOGY # * 272129 connective tissue diseasectd 2009 1 2013 4 CTD CTD CTD CTD 780 438 56. 2% 195 25. 0% 85 10. 9% 44 5. 6% SSc 18 2. 3% 286 36. 7% 8. 7% P < 0.
CHINESE JOURNAL OF ALLERGY & CLINICAL IMMUNOLOGY # * 272129 connective tissue diseasectd 2009 1 2013 4 CTD CTD CTD CTD 780 438 56. 2% 195 25. 0% 85 10. 9% 44 5. 6% SSc 18 2. 3% 286 36. 7% 8. 7% P < 0.
OncoNext TM Risk-Oncoscreening
 OncoNext TM Risk-Oncoscreening Το OncoNext TM Risk-Oncoscreening, επιτρέπει μία πολλαπλή γενετική ανάλυση που έχει στόχο την αξιολόγηση της προδιάθεσης για διάφορους τύπους κληρονομικών καρκίνων, όπως:
OncoNext TM Risk-Oncoscreening Το OncoNext TM Risk-Oncoscreening, επιτρέπει μία πολλαπλή γενετική ανάλυση που έχει στόχο την αξιολόγηση της προδιάθεσης για διάφορους τύπους κληρονομικών καρκίνων, όπως:
3.4 SUM AND DIFFERENCE FORMULAS. NOTE: cos(α+β) cos α + cos β cos(α-β) cos α -cos β
 3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
3.4 SUM AND DIFFERENCE FORMULAS Page Theorem cos(αβ cos α cos β -sin α cos(α-β cos α cos β sin α NOTE: cos(αβ cos α cos β cos(α-β cos α -cos β Proof of cos(α-β cos α cos β sin α Let s use a unit circle
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ
 ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ Μαυρόγιαννη Γεωργία 1, Καραντζούλα Ευαγγελία 2, Τουλιά Γεωργία 3, Κυριακόπουλος Βασίλης 4, Καδδά Όλγα 5 EΡΕΥΝΑ
ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ Μαυρόγιαννη Γεωργία 1, Καραντζούλα Ευαγγελία 2, Τουλιά Γεωργία 3, Κυριακόπουλος Βασίλης 4, Καδδά Όλγα 5 EΡΕΥΝΑ
Surface Mount Multilayer Chip Capacitors for Commodity Solutions
 Surface Mount Multilayer Chip Capacitors for Commodity Solutions Below tables are test procedures and requirements unless specified in detail datasheet. 1) Visual and mechanical 2) Capacitance 3) Q/DF
Surface Mount Multilayer Chip Capacitors for Commodity Solutions Below tables are test procedures and requirements unless specified in detail datasheet. 1) Visual and mechanical 2) Capacitance 3) Q/DF
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models
 Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
FORMULAS FOR STATISTICS 1
 FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT
 Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks
 98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
ΑΤΣΟΑΝΟΗ ΠΑΓΚΡΕΑΣΙΣΙΔΑ. ηέξγηνο Γειαθίδεο Γαζηξεληεξνιόγνο
 ΑΤΣΟΑΝΟΗ ΠΑΓΚΡΕΑΣΙΣΙΔΑ ηέξγηνο Γειαθίδεο Γαζηξεληεξνιόγνο Ο νκηιεηήο δελ έρεη νπνηαδήπνηε ζύγθξνπζε ζπκθεξόλησλ κε θαξκαθεπηηθέο εηαηξείεορνξεγνύο ηνπ ζπλεδξίνπ ΑΤΣΟΑΝΟΗ ΠΑΓΚΡΕΑΣΙΣΙΔΑ: νξηζκόο Η ΑΠ είλαη
ΑΤΣΟΑΝΟΗ ΠΑΓΚΡΕΑΣΙΣΙΔΑ ηέξγηνο Γειαθίδεο Γαζηξεληεξνιόγνο Ο νκηιεηήο δελ έρεη νπνηαδήπνηε ζύγθξνπζε ζπκθεξόλησλ κε θαξκαθεπηηθέο εηαηξείεορνξεγνύο ηνπ ζπλεδξίνπ ΑΤΣΟΑΝΟΗ ΠΑΓΚΡΕΑΣΙΣΙΔΑ: νξηζκόο Η ΑΠ είλαη
NMBTC.COM /
 Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
GDF-15 is a predictor of cardiovascular events in patients presenting with suspicion of ACS
 GDF-15 is a predictor of cardiovascular events in patients presenting with suspicion of ACS Δρ Στέργιος Z. Τζήκας 30.10.2015 36 ο Διεθνές Καρδιολογικό Συνέδριο, Θεσσαλονίκη Σύγκρουση συμφερόντων Δεν έχω
GDF-15 is a predictor of cardiovascular events in patients presenting with suspicion of ACS Δρ Στέργιος Z. Τζήκας 30.10.2015 36 ο Διεθνές Καρδιολογικό Συνέδριο, Θεσσαλονίκη Σύγκρουση συμφερόντων Δεν έχω
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια (MIS 365280)
 «Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
«Ειδικό πρόγραμμα ελέγχου για τον ιό του Δυτικού Νείλου και την ελονοσία, ενίσχυση της επιτήρησης στην ελληνική επικράτεια» Παραδοτέο Π1.36 Έκδοση ενημερωτικών φυλλαδίων Υπεύθυνος φορέας: Κέντρο Ελέγχου
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 651 APPENDIX B. BIBLIOGRAPHY 677 APPENDIX C. ANSWERS TO SELECTED EXERCISES 679
 APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
APPENDICES APPENDIX A. STATISTICAL TABLES AND CHARTS 1 Table I Summary of Common Probability Distributions 2 Table II Cumulative Standard Normal Distribution Table III Percentage Points, 2 of the Chi-Squared
