|
|
|
- Θεοφιλά Γερμανού
- 6 χρόνια πριν
- Προβολές:
Transcript
1
2
3
4
5
6
7 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru Saini. Supplementary Table S1: List of primers used in the present study 18S rrna FP 18S rrna RP PGC-1α FP PGC-1α RP PPARα FP PPARα RP LXRα FP LXRα RP LXRβ FP LXRβ RP SREBP2 FP SREBP2 RP SREBP1 FP SREBP1 RP HMGCS1 FP HMGCS1 RP IDI1 FP IDI1 RP LDLR FP LDLR RP ABCA1 FP ABCA1 RP ABCG1 FP ABCG1 RP ABCG5 FP ABCG5 RP NR1l3 FP NR1l3 RP CYP2B6 FP CYP2B6 RP ACACA FP ACACA RP FASN FP FASN RP HMGCR FP HMGCR RP CYP27B1 FP CYP27B1 RP GTAACCCGTTGAACCCCATT CCATCCAATCGGTAGTAGCG TGCCCT GGATTGTTGACATGA TTTGTCAGGCTGGGGGTAGG GGCGAGGATAGTTCTGGAAGC CACAGGATAAGTCACCGAGGAG GTTATAACCGGGAAGACTTTGCCA GCCTCTCTACCTGGAGCTGGT CGTGGACTTCGCTAAGCAAGTG GGTGGAAGTCGTCCTTGCTGTAGG AGGAGAACATGGTGCTGA TAAAGGAGAGGCACAGGA GCAAGGCCATCGACTACATT GGTCAGTGTGTCCTCCACCT TCCCACTCCAAATGATGACA CTTCAGGTTCTGCTGCTGTG GCATCGAGCTTTTAGTGTCTTCT GGCTGGATTGCTTAATGGATGA GTCTTGGCACTGGAACTCGT CTGGAAATTGCGCTGGAC GCACTGAGGAAGATGCTGAAA AGTTCCTGGAAGGTCTTGTTCAC CAGGAAGATTAGACACTGTGG GAAAGGGGAATGGAGAGAAGA ACCCAAAGCAAGGAACGGGAA CAGCGTTCAGCATGCCTGTGT AGTTGCACAGGTGTTTGCTG GTGCTTAGATGCTGGCATGA ATGGGGCACTGAAAAAGACTGA AGAGGCGGGGACACTGAATGAC AATCTTGAGGGCTAGGTCTTTCTGGA CCAGAGGTTGGGCCAAGGGA CAGAGTCGGAGAACTTGCAG GGAGGCATCAAACCTAGACAG TCGGTGGCCTCTAGTGAGAT TGTCCCCACTATGACTTCCC TGGCCCAGATCCTAACACATTT GTCCGGGTCTTGGGTCTAACT
8 Supplementary Table S2: The list of 1418 differentially expressed genes after overexpression of hsa-mir-195 in MCF7 cells AAMP AP4E BBS C15ORF AARSD APBA BCKDK C15ORF AATF APP BCL11A C16ORF ABHD14A AQP BCL C16ORF ACBD ARC BCYRN C16ORF ACD ARID3B BEX C16ORF ACIN ARL BIRC C16ORF ACSM ARL BMI C16ORF ACTA ARL BNC C17ORF ACTR ARL6IP BNIP3L C19ORF ADAL ARMC BP C19ORF ADAMTS ARPC1B BRAF C19ORF ADAP ARSK BRPF C19ORF ADCK ARV BSN C19ORF ADH ASB BTF3L C1ORF ADORA2B ASB BUD C1ORF ADRM ASCC BUD C1ORF AEN ASGR C10ORF C1ORF AGBL ATAD3A C10ORF C1QBP AHI ATF7IP C10ORF C1RL AHSA ATG2A C10ORF C20ORF AIM1L ATL C10ORF C2ORF AKR1C ATL C10ORF C2ORF AKT1S ATP13A C11ORF C2ORF ALDH1A ATP1B C11ORF C2ORF ALDOC ATP5L C11ORF C3ORF ALG1L ATP6V1B C12ORF C3ORF ALPL ATPBD1B C12ORF C3ORF AMOT ATRIP C13ORF C3ORF ANGEL ATXN C13ORF C3ORF ANKHD AUH C14ORF C4ORF ANKRD AUTS C14ORF C4ORF ANKRD AXUD C14ORF C5ORF ANKRD B3GALNT C14ORF C5ORF ANKRD B3GALT C14ORF C5ORF ANKRD B3GALTL C14ORF C5ORF ANKRD B9D C14ORF C5ORF ANKRD BAPX C14ORF C6ORF AP1S BAT C14ORF C6ORF AP3S BBS C15ORF C6ORF C6ORF CCNH CLK DCAF C6ORF CCRK CLN DCP1A C6ORF CCRN4L CMAS DCP
9 C6ORF CCT CNIH DDB C6ORF CCT CNOT DDTL C6ORF CD CNOT DDX C7ORF CD CNPY DDX C7ORF CD CNTNAP DDX C9ORF CDC COG DDX C9ORF CDC25C COL13A DDX C9ORF CDC42SE COL5A DEPDC C9ORF CDC5L COMMD DEXI C9ORF CDCA COPA DFNA C9ORF CDS COPS7B DHDH C9ORF CELSR COQ10A DHRS C9ORF CENPK COQ DHX CA CENPT COQ DIS3L CACNA2D CEP CORO1A DKFZP CADM CEP CP DKK CAMK2N CEP CPE DLD CASC CGGBP CPLX DLST CASC CGNL CPVL DLX CASK CHCHD CREG DMWD CAV CHD CRIP DNAH CBL CHD CRYZ DNAJB CBR CHERP CSPG DNAJC CCDC109B CHFR CTDP DNASE CCDC CHMP1A CTGLF DNASE CCDC CHST CTSC DOCK CCDC CHST CTSL DOCK CCDC CHST CTSL DOHH CCDC CHSY CTXN DPF CCDC CHURC CXCR DPP CCDC CISD CYP1B DPPA CCDC CIZ CYP26B DPYSL CCDC CKMT1A CYP27B DPYSL CCDC CLCN CYP3A DRAP CCHCR CLCN CYP4V DRG CCNB CLDN DAPK DSCR CCND CLEC4A DAXX DSTYK DUS3L ETFDH FGFBP GHR DUSP ETV FKBP GLCE DUSP EWSR FKBP GLI DUSP EXOC FLAD GLIPR DUT EXOSC FLJ GLTPD DVL EXTL FLJ GMDS DYNLRB F FLJ GNA E2F FADS FLJ GNG E2F FAM100B FLJ GNGT
10 E4F FAM105A FLJ GNL EBPL FAM111A FLJ GNS ECH FAM122B FLJ GOLGB ECSIT FAM133B FLJ GOLSYN EDA2R FAM136A FMO GORASP EDC FAM13B FNBP GPC EEA FAM158A FOXA GPC EEF1A FAM162B FOXC GPER EFNB FAM174B FOXQ GPKOW EFTUD FAM188B FRAG GPM6B EFTUD FAM193A FRMD4A GPR EIF2A FAM27A FTH GPR172A EIF3B FAM39E FXC GPR EIF3D FAM3A FXR GPR EIF3J FAM3C FYCO GPS EIF4G FAM40A FZD GRN ELAC FAM73B GABBR GRWD ELMOD FAM80B GAD GSTA ENDOD FAM84B GADD45B GSTA ENO FAM98A GADD45G GSTM EPC FANCI GALK GTF2F EPHA FARS GALNT GUSBL EPHB FARSA GALR HADHA ERAL FBRS GAS HADHB ERAP FBXL GAS HAS2AS ERCC FBXL GBF HBEGF ERCC FBXL GBGT HBQ ERF FBXL GBP HCCA ERP FBXW GCA HCFC ERRFI FDX1L GCAT HCFC ESD FGD GGT HDHD1A HELZ IGSF KCTD LINCR HESX IGSF KDM5B LIPA HEY IL28RA KDM6B LITAF HEY IL KIAA LMBRD HGS ILKAP KIAA LMF HIST1H1C IMPA KIAA LMNB HIST1H2AM INO KIAA LMTK HIST1H4H INO80D KIAA LOC HIST1H4K INPP5E KIAA LOC HIST2H2AA INPP5K KIAA LOC HIST2H2AC INSIG KIAA LOC HKR INTS KIAA LOC HLA-DMB IP6K KIAA LOC HLA-DPA IPO KIAA LOC HLA-DRB IPO KIAA LOC
11 HMGN IQCC KIAA LOC HMHA IQCK KIAA LOC HMOX IRAK KIAA LOC HNRNPA IRS KIF LOC HNRPA1L IRX KIF2C LOC HNRPA1P IRX KIF LOC HNRPDL IRX KIT LOC HOXB ISG KLF LOC HP1BP ISG20L KLF LOC HPRT ISL KLHL LOC HRAS ISOC KLHL LOC HSBP ITGAV KPNA LOC HSD17B IVD KPNA LOC HSD17B IWS KRCC LOC HSPB JAKMIP KRI LOC HSPBL JARID1A LACTB LOC IARS JHDM1D LAMC LOC ID JMJD1A LANCL LOC IER JMJD2C LARP LOC IER5L JUNB LEMD LOC IFI KAL LGI LOC IFI KBTBD LHPP LOC IFIT KCNJ LHX LOC IFT KCNK LIME LOC IGFBP KCNMB LIN LOC LOC LOC LRRC MKI LOC LOC LRRC MKX LOC LOC LTA MLL LOC LOC LTA4H MMD LOC LOC LTBP MOBKL2B LOC LOC LTBR MORF4L LOC LOC LXN MORG LOC LOC LYG MPP LOC LOC LZTFL MPP LOC LOC MAD1L MRPL LOC LOC MAF MRPL LOC LOC MAGOHB MRPL LOC LOC MAL MRPL LOC LOC MAOA MRPS LOC LOC MAP1S MRPS LOC LOC MAP2K MRPS LOC LOC MAP3K MRPS LOC LOC MAP3K MTCH LOC LOC MAP3K MTHFD2L LOC LOC MAP7D MTHFR LOC LOC MAPK MTIF
12 LOC LOC MCCC MTRF1L LOC LOC MCL MUC LOC LOC MCM MUM LOC LOC MCM MURC LOC LOC MDM MUS LOC LOC MEA MYBBP1A LOC LOC MED MYC LOC LOC MED MYCBP LOC LOC METAP MYH LOC LOC METTL MYL LOC LOC METTL MYO3A LOC LOC MFSD NAB LOC LPAR MGC NADSYN LOC LPL MGC NAIF LOC LPPR MGC NAPB LOC LPXN MGC NAPG LOC LRIG MGMT NARFL LOC LRIG MICALL NAT LOC LRP5L MIRLET7D NCAPD NCAPG NUP PDX PPAN NCKAP NUP PDXDC PPAP2A NCOA OAZ PEA PPIA NDUFB ODF PECR PPIC NEK OFD PELP PPP1CB NEK OIP PEX PPP1R12C NELF OKL PGBD PPP1R14C NELL OSAP PGD PPP1R15A NETO OSBPL1A PGS PPP2R5B NEU OSGEPL PHC PPP3CB NEUROG OVGP PHF21B PPPDE NEXN OXR PHIP PPRC NFKB OXSR PHLDA PQLC NFKBIA OXTR PIAS PRCP NFKBIB PA2G PIGF PRDM NFKBIL PACSIN PIGV PRDX NFRKB PAF PIGY PRKAB NHP2L PAFAH1B PIK3IP PRKCABP NIF3L PAFAH1B PIK3R PRKCH NKD PAM PIP4K2A PRKRIP NKTR PAPD PIR PRMT NLE PAQR PISD PROS NOC2L PARM PLA2G4C PRPF NOC3L PARP PLA2G PRPF NOL PATL PLAC8L PRR NOL PAX PLD PRR NOP PBK PLEKHF PRRG
13 NOP PBX PLEKHM PRRG NPIP PBX PLS PRRX NPY PCGF PLSCR PRSS NR1H PCMT PMS2L PRSS NR2F PCMTD PNMA PRSS NSUN PCOLCE POFUT PSAP NT5C PCSK1N POLA PSEN NUBP PCSK POLE PSMD NUCB PDCD2L POLM PTBP NUDT PDGFD POLR2A PTCD NUDT PDGFRL POLR3A PTCH NUDT16L PDHB POLR3D PTDSS NUDT PDIA PON PTGER PTGER RHBDD RRN SETD PUSL RHBDD RRP SETD PVRL RHBDL RRP SETX PYCR RHOT RRP7A SF PYGB RIC8B RSC1A SFRP QARS RLF RSHL SFRS QPRT RN5S RSPH SFRS QTRT RNF RSRC SFRS RAB RNF RTN4IP SFRS17A RAB11A RNF RUFY SFRS RAB RNF RUSC SFRS RAB6B RNF S100A SFXN RAB7L RNF5P SAC3D SGK RAB9B RNMT SAFB SH2B RAD23B RNU SAFB SH3BP5L RAD9A RNU SAMD SHARPIN RAGE RNU1A SAMD SIAH RALA RNU1F SAP SIGMAR RALB RNU1G SAPS SIRT RALGAPB RNU SAT SIX RAP2A RNU SBDS SIX RAPGEF RNU6ATAC SCAMP SKAP RASSF RNU SCHIP SLC12A RASSF RNY SCOC SLC19A RBBP RNY SCYL SLC1A RBKS RP11-529I SDCCAG SLC22A RBL RP9P SDHAP SLC30A RBM RPA SDHD SLC35A RBM RPAP SEC22A SLC35B RBM RPF SEC SLC35D RBM RPL13A SELENBP SLC36A RBM RPL SELM SLC37A RBMS RPL SELO SLC38A
14 RBMX RPL7A SEMA3A SLC3A RC3H RPN SEMA6A SLC44A RDH RPS SEPW SLC47A RECQL RPS SERPINB SLCO4C REEP RPS26L SERPINE SLIT RFWD RPS6KA SERTAD SLITRK RFX RPUSD SETD1A SMPD SMS ST6GALNAC TMED TRMT61A SMYD STAG TMED TRMU SNAPC STK11IP TMEFF TRNAU1AP SNCA STK TMEM TSHZ SNHG STK TMEM TSHZ SNHG STRADB TMEM TSPAN SNORA STRN TMEM TSPAN SNORA STS TMEM TSPAN SNORD STX1A TMEM TSR SNORD14A STX TMEM TSSC SNORD SUCLA TMEM TSTA SNORD3A SUCLG TMEM TTC SNORD3C SUMO TMEM TTC SNORD3D SUMO TMEM41B TTC SNORD SUPT5H TMEM TTC SNTB SUPT6H TMEM TTLL SNUPN SURF TMEM TTLL SNX SURF TMEM90B TUBB SOX SUSD TMSB15A TUBB2B SOX TAF TMX TUFT SPAG TAF TNFAIP TUSC SPATA2L TAF9L TNFRSF12A TWIST SPINK TAP TNIP TXNDC SPINK TAPT TNPO TXNL4B SPON TATDN TOM1L TYW1B SPRY TBC1D22B TOP3B U2AF1L SPRYD TBL TOPBP UAP1L SPTLC TDG TOR1AIP UBA SQSTM TEX TP53BP UBAP SRI TFPT TPX UBE2J SRM TGFBR TRAFD UBE2N SRPX THADA TRIB UBL SRRM THEM TRIM UBQLN SSBP THOP TRIM UBR SSH THUMPD TRIM UBR SSR TIMM TRIM UCRC SSX2IP TIMP TRIM UFSP ST TJAP TRIP UIMC ST3GAL TLE TRIT UMPS
15 ST3GAL TLE TRMT UNC84A UPLP ZFAND2A ZNF USP USO ZFAND ZNF USP UST ZMAT ZNF XPC UVRAG ZMIZ ZNF XRCC VASN ZNF ZNF XRCC VGF ZNF ZNF XRCC VPS ZNF ZNF YEATS VPS33A ZNF ZNF ZADH VPS33B ZNF ZNF ZBTB VPS37B ZNF ZNF ZBTB WARS ZNF ZNF ZBTB WASF ZNF ZNF ZBTB WASH2P ZNF ZNF ZBTB WBSCR ZNF ZNF ZBTB WDR ZNF ZNHIT ZBTB WDR ZNF ZNRF ZC3H WDR ZNF ZP ZC3H WDR ZNF ZRSR ZDHHC WDR ZNF ZSCAN ZFP WDR ZNF ZSCAN ZFP WHAMM ZNF ZZZ ZNF ZNF WNT5A WNT5A ZNF ZFPM ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF ZNF
16 Supplementary Table S3: The list of 428 differentially expressed genes after depletion of hsa-mir-195 in MCF7 cells AEN C20ORF CXORF GTF3C AKR1C C20ORF CYP27B HAVCR ALPL C21ORF CYP4V HESX ANGEL C4ORF DAPK HIST1H2AC ANKRD C6ORF DCP HIST1H4K ANKRD C6ORF DDB HIST3H2A APBA C6ORF DDX HJURP ARC C6ORF DKFZP686K HLA-E ARF C7ORF DNTTIP HSD17B ARMC C7ORF DPP HSPB ASGR C8ORF DUS3L HTRA ATAD CBR DUSP IER ATAD3A CCDC DYNLL IGSF ATL CCDC E2F IL ATP5G CCDC ECT IL ATP5J CCDC EDC IRF AURKAIP CCL EIF2S ISG20L AVL CCT EIF3A KIAA AYP1P CD ELP KIAA BCL CDC25A ERF KIF2C BLOC1S CDCA ERRFI KIF4A BRI CDCA FAM133B KIFC BRI3P CDCA FAM158A KRT BTN3A CENPBD FAM3A LACTB BUB CENPE FAM46C LAGE BUB1B CENPN FAM98A LENG C10ORF CENPQ FAU LGALS C10ORF CHD FDPS LINS C11ORF CHERP FDX1L LOC C12ORF CHMP1A FHOD LOC C12ORF CHP FLJ LOC C14ORF CIR FLJ LOC C14ORF CKS FRG LOC C15ORF CMC GADD45A LOC C15ORF CORO GMPR LOC C16ORF COX6B GNG LOC C19ORF COX7B GNGT LOC C19ORF CPOX GPATCH LOC C1ORF CXCR GPR172A LOC LOC LOC MGC ODF LOC LOC MGC ORC1L LOC LOC MICB OSAP LOC LOC MIR PATL LOC LOC MIR PELP LOC LOC MKL PFAS LOC LOC MKX PHF21B LOC LOC MPHOSPH PHLDA LOC LOC MRPL PHPT LOC LOC MRPL PIBF LOC LOC MRPL PISD LOC LOC MRPL PLA2G4C LOC LOC MRPS PMS2L LOC LOC MT1A PNMA LOC LOC MT1F POMT LOC LOC MT1X PPP1R12C LOC LOC MT2A PRMT LOC LOC MTE PTTG3P
17 LOC LOC MTIF RALGAPB LOC LOC MTMR RASSF LOC LOC MVD RBM LOC LOC MYBBP1A RBM LOC LOC MYBL RDH LOC LOC NADSYN RECQL LOC LOC NCAPD RFWD LOC LOC NCAPD RHOT LOC LOC NCAPG RIOK LOC LOC NDC RN5S LOC LOC NDUFA RNF LOC LOC NDUFB RNU LOC LOC NDUFB RNU LOC LOC NEIL RNU1A LOC LSMD NEK RNU1F LOC LTBP NEU RNU1G LOC MAP2K NFKB RNU4ATAC LOC MAP3K NFKBIA RNU LOC MAP7D NFKBIB RNU LOC MCM NIF3L RNU LOC METRN NPM RNY LOC METTL11A NRAS ROMO RP9P SHROOM STX TTLL RPL12P SKIV2L SUGT TUBD RPL23AP SLC2A SYT TUT RPN SLC35A TACC TXNDC RPRML SLC35B TAF UBA RPS SLC36A TAP UBAP RPS SLC5A TBL UBL RPS SLU THAP UBLCP RPS26L SMNDC THOP UHRF RPS SNAP TIMM8A USP RRP SNHG TMEM UST RRP SNHG TNFAIP UTP14A RRP7A SNORA TNFRSF VGF RSRC SNORA59B TNIP VPS33B SAP SNORD3A TOMM20L VPS4A SBDS SNORD3D TOMM VTI1B SCNM SNORD TPX WDR SDC SNORD TRAF YES SDHD SNRPF TRIM ZADH SEC61B SNRPG TRIP ZNF SELM SPATA5L TRIT ZNF SEPW SPG3A TRMT61A ZNF SERTAD SQSTM TSHZ ZNF SH3BP SRM TSHZ ZNF SHFM STIL TTK ZNF ZNF ZNF ZNF
18 Supplementary Table S4: List of differentially expressed genes showing significant fold difference of expression levels in case of hsa-mir-195 overexpression and inhibition in MCF7 cells Gene name Fold change after mir- 195 overexpression Fold change after mir- 195 inhibition Difference in Fold change PLA2G4C PHF21B RNY RNU SNHG RNU1G SNORD3D GNGT TRMT61A RNU LOC ARC ANKRD RNU AEN RNU1A TRIT HIST1H4K CCDC VGF RN5S RASSF SERTAD PMS2L DAPK RRP TTLL DUSP KIAA SBDS DUS3L SNORD3A THOP C10ORF LOC HSD17B SDHD
19 Supplementary Table S5: The list of networks generated during Ingenuity pathway analysis using the differentially expressed genes obtained after hsa-mir-195 over-expression in MCF7 cells Molecules in Network Score Focus molecules ACIN1, AKR1C3, Akt, AKT1S1, ANGEL1, ARL1, ARMC6, ATP5L, CADM1, CHFR, CREG1, DNA-directed RNA polymerase, EFTUD2, FOXC1, INPP5E, MIRLET7D (includes EG:406886), MPP3, NEXN, NOC2L, PDXDC1, PHLDA3, POLR3A, POLR3D, PROS1, QARS, RNF115, RPAP1, RPN2, SAP18, SFXN1, SLC3A2, STK36, TGFBR3, ZC3H18, ZZZ3 BTF3L4, CTDP1, DDX56, DRAP1, ERF, FANCI, GAS1, GNL2, GTF2F1, HELZ, HLA-DR, HLA-DRA, HLA-DRB1, Holo RNA polymerase II, MRPS5, MYBBP1A, MYC, MYCBP2, NELF, PAF1, POLR2A, RAGE, RNA polymerase II, RPL15, RRP12, SAFB, SETD2, SLC27A2, SPAG7, SSBP2, SUPT5H, SUPT6H, TAF10, TAF9B, UST ATL2, AUH, AUTS2 (includes EG:26053), C12ORF44, CCND2, COPA, CTSC, DKK3, ECH1, enoyl-coa hydratase, ERP29, EWSR1, FGFBP1, GBP2, HADHA, HADHB, IgG, Immunoglobulin, Interferon alpha, MED4, MED14, Mediator, NKTR, NLE1, PAFAH1B3, PATL1, PDIA6, PRKAB1, RSRC2, SERTAD1, TRIM28, UBQLN4, WBSCR22, ZNF205, ZNRF3 AATF, BBS2, C15ORF23, C5ORF13, COMMD1 (includes EG:150684), CORO1A, DCP2, DCP1A, E2F7, EDC3, GNGT1, GPC4, IRAK2, LITAF, MHC CLASS I (family), MPP6, MRPL38, NFKB1, NFkB (complex), NFkB (family), NFKBIB, NOP14, Proinflammatory Cytokine, Serine Protease, SIAH2, SQSTM1, SUMO3, THOP1, TNFRSF12A, TNIP1, TRAFD1, TUBB2B, Ubiquitin, ZFP36, ZNF202 ACTA1, Alp, ALPL, APBA3, APP, ARL6IP5, BCKDK, BIRC6, C14ORF147, Caspase, CBR4, DNAH1, DUSP14, FSH, GCA, GPS2, HSD17B8, IARS2, IGFBP5, JUNB, KDM5B, Lh, LTBP2, PBK, PPAP2A, PPP2R5B, PSEN2, PSMD3, SMPD2, SPON1, SRI, SUCLA2, Tgf beta, VGF, ZNF773 AP1S2, Ap2 alpha, ATPase, C3ORF1, CBL, CDK20, DLX1, DUSP5, EPHA4, GHR, GPC3, growth factor receptor, HGS, IER5L, ILKAP, INO80, INO80D, IPO13, NAPB, p85 (pik3r), PAX6, Pdgf, PDGF BB, PDGFD, PDX1 (includes EG:3651), PELP1, PICK1, Pkc(s), PLC gamma, PPP1R15A, SOX2, TIMP3, TRIM11, VPS37B, WNT5A ADAP2, ANKRD1, CHD8, ENaC, ERK1/2, FKBP11, GRN, HERP, HEY1, HEY2, HIST1H1C, KLF13, METAP2, Notch, P- TEFb, peptidase, peptidylprolyl isomerase, PPI, PPIA (includes EG:5478), PPIC, PRDX6, PRSS8, RNF10, RNU1-4, S100A4, Secretase gamma, SEMA3A, SERPINE2, SH2B3, SLC12A4, SNAPC4, SNCA, TOP3B, VASN, ZFPM1 (includes EG:161882) APC, CCDC76, CCNB1, CDC5L, Cyclin A, Cyclin B, Cyclin E, DAPK3, DAXX, E2f, E2F5, E4F1, Hdac, ID2, KIAA0240, LIN37, MAP1S, MCM2, MCM5, MDM4, NOP2, PCSK7, PCSK1N, PEA15, PRPF19, Ras, RASSF1, Rb, RBBP9, RBL2, RBMX, TSHZ3, Xrcc, XRCC1, XRCC3 (includes EG:7517) ACTR6, ADH5 (includes EG:128), Ahr-aryl hydrocarbon-arnt, alcohol dehydrogenase, BCL3, BCL11A, COL13A1, Ctbp, CYP1B1, CYP27B1, CYP3A5, DHRS2 (includes EG:10202), DUSP8, EDA2R, EPC1, ERRFI1, glutathione transferase, GST, GSTA1, GSTA4, GSTM2, ISL1, Jnk, NR1H3, NR2F1, PECR, PIR, PXR ligand-pxr-retinoic acid-rxrα, RAB7L1, Rxr, SERPINB1, TIP60, TRIP11, VitaminD3-VDR-RXR, YEATS4 ADAMTS1, ASB9, BNIP3L, C1ORF182, CCT4, COL5A1, collagen, Collagen(s), Cpla2, CTSL1, EXOC3, F12, FAM40A, FTH1, GADD45B, GADD45G, Hsp27, Hsp70, HSPB8, IL1, IL32, KLF9, NCOA5, P38 MAPK, PLA2, PLA2G7, PLA2G4C, Pld, PLSCR3, RALA, RAP2A, RAPGEF6, RPS6KA4, Sapk, STRN3 ASCC2, CD19, EEA1, ELAC2, Gamma tubulin, HMOX1, IFIT2, IFN Beta, IL12 (complex), IL28RA, IRG, ISG15, JAK, KDM3A, KDM5A, KIT, LDL, LIPA, LT, LTA, LTBR, MKI67, NAPG, PI3K (complex), PIK3R4, PON2, Rab5, RUFY1, SLIT2, STAT5a/b, TNFAIP3, TRAF, UVRAG, VAV, VPS18 ATRIP, Cbp/p300, Cofilin, DDB2, DPYSL2, ERCC2, ERCC3, ERK, GRWD1, Importin beta, IPO8, KPNA1, KPNA4, POLE, RAD23B, RAD9A, RNA polymerase I, Rock, RPA, RPA4, RRN3, SAA, Smad2/3-Smad4, SNUPN, SOX9, TDG, TFIIA, TFIIF, Top functions Cellular Movement, Reproductive System Development and Function, Cardiovascular Disease Gene Expression, RNA Post- Transcriptional Modification, Antigen Presentation Genetic Disorder, Metabolic Disease, Lipid Metabolism Hematological Disease, Organismal Injury and Abnormalities, Gene Expression Lipid Metabolism, Molecular Transport, Small Molecule Biochemistry Cellular Development, Cellular Growth and Proliferation, Cell Morphology Cellular Growth and Proliferation, Nervous System Development and Function, Tissue Development Cell Cycle, DNA Replication, Recombination, and Repair, Connective Tissue Development and Function Drug Metabolism, Glutathione Depletion In Liver, Lipid Metabolism Cell Signaling, Carbohydrate Metabolism, Lipid Metabolism Cardiovascular System Development and Function, Lymphoid Tissue Structure and Development, Organismal Development DNA Replication, Recombination, and Repair, Nucleic Acid Metabolism, Small Molecule Biochemistry
20 Molecules in Network Score Focus molecules TFIIH, TOPBP1, TP53BP1, UBR2, UBR5, XPC, ZBTB17 ARPC1B, ATL3, C11ORF75, C16ORF48, C6ORF72, DHX8, DMWD, DPPA2, DVL3, DVL2 (includes EG:1856), ENDOD1, FXR2, MAPK1IP1L, MIR124, MOBKL2B, NOC3L, NT5C2, PLOD3, PRPF3, RAD51AP1, SDF4, SLC30A7, SRM, SUCLG2, TCEB3B, THAP1, TRIM29, TSR2, USP5, USP39, ZBTB8A, ZNF165, ZNF250, ZNF263, ZNF446 Ap1, ARL2, Arp2/3, ASGR1, Calmodulin, CaMKII, CEP250, Ck2, Creb, CSPG5, EIF3, EIF2A, EIF3B, EIF3D, EIF3J, EIF4G3, F Actin, GAD1, MGMT, MICALL1, MYH3, MYL5, Myosin, PACSIN1, PIP4K2A, PLS1, POLM, PP1 protein complex group, PP1-C, PPP1CB, PPP1R14C, SIX1, SSR1, Tubulin, XRCC5 ACVRL1, ADAM12, AP3S1, BUD31 (includes EG:8896), C6ORF130, CNTNAP2, CTR9, DFNA5, E2F5, EEA1, EGLN1, ENO1, G3BP1, HSD17B6, KDM6B, KRI1, LDB1, NKX3-2, NOL6, NUP214, PAPPA, PDLIM5, PIAS3, PIGF, RUNX3, SMAD2, SNIP1, SSBP4, SURF6, TGFB1, TGFBR3, TLE4, TPM3, VASN, ZMIZ1 ADCY, ADORA2B, CELSR3, CXCR7, DOCK2, Focal adhesion kinase, FZD4, G protein, GABBR2, GALR2, Gpcr, GPER, GPR37, GPR64, GPR160, GPR172A, Gs-coupled receptor, Gsk3, HBEGF, IL12 (family), LPAR3, LPXN, Mmp, NPY, Pias, PIAS3, Pka, PLC, PTGER1, PTGER2, Rac, SRC, STAT, ZNF426, ZP3 ACBD6, C11ORF54, CCDC45, CHERP, GMDS, HNF1A, HNF4A, ISOC1, LSM3, MCCC1, RBKS, RSPH3, RTN4IP1, SFRS8, SMAD3, TTC25, TUFT1, TXNL4B, WARS2, WBP4, ZNF277 ARRB1, ATAD3A, BAT1, BAT5, BCLAF1, BUD13, CCDC93, CCL14, CHCHD3, CHD1 (includes EG:1105), CWC22, DDX39, DHRS2 (includes EG:10202), DHX16, FRMD4A, GPKOW, GRK6, HNRNPA1, HNRNPA1L2, IGF2BP2, KIN, LARP7 (includes EG:51574), LMNB2, MRPS27, PIP, PPIA (includes EG:5478), PRSS23, RBMX2, RECQL4, SAFB, STOM, THOC4, TMEM222, TRAF3IP1, ZBTB43 AHSA2 (includes EG:130872), APPBP2, ARID3B, BCL3, BDP1, BNC2, BRF1, CMAS, CREG1, DNAH1, DNASE1, F2R, Gcn5l, HSP90AA1, IKZF1, IRF1, KDM5A, MDM4, MED4, NR3C1, PI4KAP2, RB1, RBMS2, RNASEN, SNAPC1, SNAPC3, SPTAN1, STRADB, TAF4B, TBL3, TBP, TNFAIP3, TRIM45, TSSC4, WDR62 C19ORF42, FAM158A, HIST1H4K (includes EG:8362), HMGN4 (includes EG:10473), HNF4A, KLHL28, LRIG2, NUDT11, ONECUT1, SDR39U1, SLC35A5, TMEM59 (includes EG:9528), TRAF6, TRMT6, TRMT61A, TTC35, TTC38, ZNF304, ZNF324, ZNF557 AKAP1, C16ORF74, C5ORF41, CDKN1A, CIZ1, DAXX, DPF1, DPP3, FEN1, FGF13, GPS2, KBTBD8, LENG8, LGALS1, LIG1, MDM4, MIR106B (includes EG:406900), MIR17 (includes EG:406952), MIR196A2, MKX, NEUROG2, PARP2, PARP6, PLK2, PNPT1, PPP3CA, RB1CC1, RIMKLB, RNF121, SMOC2, SOX9, TP73, TTLL5, UBL3, XRCC1 CALM2, CCDC66, CENPT, CFL1, CLCN6, CTU2, DDX6, DHX29, FBXO33, FKBP8, GLCE, GNG4, KIAA1683, LRRC59, LSM12, MIR205 (includes EG:406988), MIR24-1 (includes EG:407012), MIR31 (includes EG:407035), MTCH2, MXI1, OFD1, PDGFRA, PHF21A, PISD, PRRX2, RAB2A, RAB5C, RAB9B, RIC8B, RNF10, RPL22, SDCCAG8, Tropomyosin, VHL, WDR55 Top functions RNA Post-Transcriptional Modification, DNA Replication, Recombination, and Repair, Metabolic Disease Protein Synthesis, DNA Replication, Recombination, and Repair, Connective Tissue Disorders Gene Expression, Cellular Development, Cellular Growth and Proliferation Cancer, Neurological Disease, Cardiovascular System Development and Function Gene Expression, Embryonic Development, Tissue Development RNA Post-Transcriptional Modification, Molecular Transport, RNA Trafficking Gene Expression, Respiratory Disease, Digestive System Development and Function Digestive System Development and Function, Hepatic System Development and Function, Organ Development DNA Replication, Recombination, and Repair, Cell Cycle, Cellular Development Tissue Morphology, Carbohydrate Metabolism, Drug Metabolism , alcohol group acceptor phosphotransferase, BRAF, CAMK2N1, ECSIT, ETV5, HRAS, IKK (complex), Ikk (family), LGALS1, MAP2K2, MAP2K1/2, MAP3K, MAP3K1, MAP3K9, MAP3K11, Mapk, MAPK13, Mek, MUC16, Nfat (family), OIP5, Pak, PP1/PP2A, PRKCH, Raf, Rap1, RC3H2, RFWD3, RPS23, Sos, TCR, TRIB1, UBA6, UBE2N CD79B, CDCA2, DAXX, DEXI, DUT (includes EG:1854), EDIL3, ELK3, ETS1, FAM3C, GOLM1, ING4, LMTK2, LTB, NEK2, NOC2L, P2RX4, PAFAH1B3, PPP1CA, PPP1CC, PPP1R2, PPP1R13B, PPP1R15A, PPP2R5C, RAD23B, REEP5, RPL13A, SCOC, SEC62, TAF1A, TMSB15A (includes EG:11013), TP53, ZAP70, ZMAT3, ZNF140, ZNF24 (includes EG:7572) ADCK1, AIF1L, AP1G1, AP3S2, AP4E1, AP4M1, Arf, ARF1, ARF5, ARF6, ARFGAP1, ARFIP2, CAV2, CEP350, CLCN7, COPG, GBF1, GOLGB1, GOSR2, HOXA9, HOXB8, IRX3, IRX6, MIR210 (includes EG:406992), oleic acid, OSBPL8, PACS1, PBX3, PBXIP1, PIK3IP1, PLEKHM2, QTRT1, RAB1A, Developmental Disorder, Genetic Disorder, Cellular Movement Cell-To-Cell Signaling and Interaction, Cellular Function and Maintenance, Cell Death Molecular Transport, Protein Trafficking, Cellular Assembly and Organization
21 TMEM35, USO1 Molecules in Network Score Focus molecules Top functions Supplementary Table S6: The list of networks generated during Ingenuity pathway analysis using the differentially expressed genes obtained after hsa-mir-195 deletion in MCF7 cells Molecules in Network Score Focus molecules Top functions ACLY, BUB1, BUB1B, CENPE, CKS1B, CSE1L, DHCR7, DUSP5, EIF2S1, EIF3A, GADD45A, IFN Beta, IFN TYPE 1, IgG, IRF1, IRG, ISG15, KRT8, LDL, MAD1L1, Mapk, NDC80, NEK2, PP1 protein complex group, PP1-C, PP2A, PPP1R1A, PRKRA, RPL5, RPS23, SLC2A3, STIL, TAP1, Vegf, YES1 ALPL, Ap1, CD70, COX6B1, COX6C, COX7B, COX8A, Cytochrome c oxidase, DNTTIP1, ELP4, Estrogen Receptor, FSH, GLRX2, Histone h4, Ifn gamma, Jnk, KIFC1, LSM7, LTBP2, MEPCE, MT1F, MYB (includes EG:4602), Pdgf, PELP1, PHLDA3, Pias, RANGAP1, Rxr, SNRPF, SNRPG, Tgf beta, TRIM26, TRIP12, TUT1, VGF alcohol group acceptor phosphotransferase, Alpha tubulin, APC, AURKA, AURKAIP1, BIRC5, BTG2, CDC25A, CKS2, Cpla2, Cyclin A, Cyclin B, Cyclin E, CYP27B1, E2f, E2F5, ECT2, ERK1/2, Gamma tubulin, Hdac, IDE, KIF2C, MAP3K8, MAPKAPK5, MCM5, MT2A, MYBL2, ORC1L, PLA2G4C, RACGAP1, RASSF1, RPA3, TPX2, TSHZ3, TTK 26s Proteasome, Akt, ASGR1, BLOC1S2, Caspase, CCL2, CDCA3, ERRFI1, GNGT1, hcg, Hsp27, Hsp70, HSPA5, IFT57, Ikb, IL32, Importin beta, NfkB1-RelA, NFKBIA, NFKBIB, NIF3L1, PTTG1, RIOK3, SAA, Sapk, SDHA, SRP19, SSR4, STIP1, TRAF2, TRIAP1, TXNDC17, Ube3, Ubiquitin, USP5 BCL3, CAMK2N1, CD3, FTL, Gm-csf, HLA-E, IER3, IKK (complex), Ikk (family), LGALS1, MAP2K2, MAP3K, Mek, MHC Class I (complex), MHC CLASS I (family), MPHOSPH6, NFKB1, NFkB (family), NfkB-RelA, NRAS, PDGF BB, PEBP1, PI3K (complex), PIK3R4, Raf, RHOB, RPN2, SERTAD1, TCR, THOP1, TNFAIP3, TNFRSF9, TNIP1, TRAF, TYRO3 ATAD3A, BTG2, CCDC92 (includes EG:80212), CDCA2, CDKN2AIP, CPOX, CTSK, EIF2S1, FGF2, HJURP, HNRNPA1L2, HTRA1, IFNB1, ISG15, MAD1L1, MKI67, MT1A, NEK2, NOC2L, PPP1CA, PPP1R10, PPP2R5C, PRKRA, RECQL4, RNU1-1, RPS28, S100A8, STOM, TACC3, TAP1, TERT, TOX4, TP53, TXNRD1, ZC3H4 ARF3, Calmodulin, CDCA5, CHMP1A, DCP2, EDC3, EDF1, ERK, F Actin, GDI1, GMPR, Histone h3, IL8, Immunoglobulin, KIAA0895, KIF23, MIR1, MSN, Nfat (family), NGF, P38 MAPK, PATL1, Pkc(s), Pld, PNMA2, Rac, RALA, Ras, Ras homolog, Rho gdi, RHOT2, SDC4, SH3BP1, SHROOM3, UST AKR1C3, ANGEL1, CCHCR1, DEFB4A, EBF3, FABP6, FRG1, IFNGR2, ME1, MIRLET7D (includes EG:406886), MRPL19, MRPL41, MVD, NCF1C, NEU1, NFKB1, NKRF, NR4A2, PMS2L3, PTEN, RETN, SMURF2, T3-TR-RXR, TAL1, TARBP2, TICAM1, TNFAIP1, TNFAIP3, TNFRSF9, TP53BP1, TRAF, TRAF4, UTP14A, VPS4A CD3EAP, CKMT1A, GTF3C2, IKBKE, KIF11, LAGE3, METTL11A, MKX, MT-ND1, MT-ND4, NADH2 dehydrogenase, NADH2 dehydrogenase (ubiquinone), NDUFA1, NDUFA3, NDUFA6, NDUFA13, NDUFB7, NDUFB9, NDUFB10, NDUFS1, NDUFS8, NUTF2, PFAS, POLR3C, RNY4, SBDS, SDHA, SDHB, SDHD, SNORD3A, SSB, TBP, TNFAIP3, TROVE2, VEGFA ANPEP, ARC, AUH, AZU1, BTN3A3, CD40LG, CSNK1D, DAPK3, DEFA1 (includes EG:1667), DYNLL2, EGF, Gelatinase, GNLY, HIST3H2A, IL15, IL32, IL1R2, Lh, MDM2, MICB, MIR152 (includes EG:406943), NFRKB, PAWR, POU2F1, RASAL2, RNF217, RNU6-1, RPS21, SLU7, THAP10, TNF, TNFRSF9, TNFSF9, TNIP1, VCL ARL6, ASGR2, BAT3, C20ORF111, CDC45, CHERP, CIAO1, CIR1, COPB2, DPM1, GNL3, GTF3C5, HDAC8, HNF4A, LSMD1, MRPL15, MRPS15, MT1X, PHPT1, POLR1C, PSMB1, RPAP1, SEC61B, SHFM1, Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair Energy Production, Cell Cycle, Cell Morphology Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair Cancer, Hematological Disease, Organismal Injury and Abnormalities Cell Cycle, Gene Expression, Cell Signaling Cellular Development, Cellular Growth and Proliferation, Nervous System Development and Function DNA Replication, Recombination, and Repair, Cellular Assembly and Organization, Post- Translational Modification Immunological Disease, Gene Expression, Infectious Disease Cancer, Neurological Disease, Genetic Disorder Inflammatory Response, Drug Metabolism, Lipid Metabolism Carbohydrate Metabolism, Nucleic Acid Metabolism, Small
22 Molecules in Network Score Focus molecules SLFN11, SNW1, SRP54, SRSF2, TROVE2, VIPAR, VPS41, VPS33B, WBP4, WBSCR22, ZNF764 Top functions Molecule Biochemistry ANKRD1, Ck2, DDB2, DNAJC7, DNAJC10, Fibrinogen, GTF3C5, HSP, Hsp90, HSPA14, HSPB6, HSPB7, HSPB8, Ifn, IL1, IL32, IL12 (complex), IL12 (family), IL18RAP, IL1F6, IL1F8, IL1F9, Interferon alpha, Mmp, NELF, NFkB (complex), NPM3, ODF1, PIBF1, RNA polymerase II, SQSTM1, SUGT1, TAF10, WDR34, XAB2 Actin, ACTR8, ALDH9A1, APEX2, ARFIP1, ATL3, C12ORF45, CDC14B, CDCA7L, DCTD, DEPDC1, ELF4, KIAA1468, KIF4A, MIR124, MIR124-1 (includes EG:406907), MTMR6, NCAPD2, NCAPG (includes EG:64151), NCAPH, NECAP2, NEK6, OSBPL8, PFDN6, PTTG1IP, RDH, RDH10, RDH11, RDH13, RUVBL2, S100A11, SECISBP2, SEPW1, SURF4, TMEM hydroxyeicosatetraenoic acid, ADAM17, APBA3, C11ORF70, C4ORF49, CAMLG, CENPN, CNTF, EGFR, Egfr-Erbb2, FHOD1, G-protein beta, GRB2, Il8r, IRS1, MAP2K5, MIR122 (includes EG:406906), MRPL36, MYCN, NDN, NUBP1, NUMBL, ONECUT1, P110, PPP1R12C, PTGER1, RIN2, RNF122, RPS5, RPS15, RRAS, TAOK3, TMEM55B, UBAP2, ZKSCAN3 AIRE, ARMC1, BCORL1, DDX23, EFTUD2, FAM178B, GNL3, HIST1H2AC, KBTBD8, MIR138-1 (includes EG:406929), MIR138-2 (includes EG:406930), MIR183 (includes EG:406959), MIR221 (includes EG:407006), MYBBP1A, MYC, NCAPD3, NCAPG2, NEIL2, NFYB, NOP56, ODF2, PCNA, PNPT1, PRPF6, PRPF8, RNMTL1, SART1, SKIV2L, SMNDC1, SNRNP200, STX16, TSHZ1, TSR2, USP39, ZNF598 ANXA1, C7ORF50, Ca2+, CALCA, CCDC86, DLST, EPO, FARSA, FAU, FOXP3, IL2, ITGA4, Laminin, METAP2, MRPS17, MTF2, PDCD2L, PINX1, POLR1A, POLR1B, POLR1C, POLR3K, PRNP, PTHLH, PTRH1, S100A11, SCNM1, sodium chloride, TBL3, TNFRSF11A, TNFSF9, TRPM4, USP11, USP25, WNT5A ACIN1, ALDOA, AP2B1, ARRB1, C1ORF35, CHD1 (includes EG:1105), CLPB, DDX39, DPP3, FAM46C (includes EG:54855), GBAS, MPND (includes EG:84954), NSF, OSBPL10, PCBP3, PGAM5, RBM17, RNPS1, SAP18, SF3A2, SF3B2, SNAP29, SNRNP70, SNRPC, SRRM2, SS18L1, STX7, STX8, TIMM8A, USP30, VAMP7, VTI1A, VTI1B, WRNIP1, ZC3H18 AEN, C17ORF61, C8ORF30A, CDC34 (includes EG:997), CHFR, COX6C, DDX58, EXOSC9, FAM176A, FDPS, heparan sulfate, hydrogen peroxide, KIAA0776, MDM4, PSMD2, PTN, RFWD3, RLIM, RNF11, RNF181, TRAF7, TRIM25, TRIM26, TRIM31, TRIT1, UBA6, UBE2D1, UBE2D2, UBE2D3 (includes EG:7323), UBE2N, UBE2V1, UBE2V2, UBE4A, UBLCP1, UHRF2 ATAD2, BDKRB1, C14ORF156, C21ORF122, CNR2, CXCR7, CYSLTR2, ESR1, FOS, Gpcr, GPR50, GPR172A, HESX1, IL9, LPAR1, LPAR2, LPAR3, MIR214 (includes EG:406996), MKL1, MKL2, NADSYN1, NR2C2, NR2C2AP, P2RY1, P2RY6, P2RY11, PISD, PTGER1, PTGER4, RPRML, RRP12, SMARCA4, SRF, STAT3, ZNF467 ABCF3, ANGEL2, AP2A1, BARD1, C19ORF44, CP110, DNAJA2, FAM158A, GBF1, GPATCH3, HIST1H4K (includes EG:8362), HNF4A, KBTBD7, KIAA1967, LENG1, LLGL1, MRPL49, PI4KB, PJA2, PNMA1, POLRMT, RAP2C, RBM26, SPATA5L1 (includes EG:79029), TOE1, TP53, TRMT6, TRMT61A, TTC35, ZNF304 ATL1, C6ORF130, CCT5, CLK2, G protein alphai, G-protein beta, G-protein gamma, GNG2, GNG3, GNG4, GNG10, GNG13, GSN, HAVCR2, ITK, LGALS9, MUC1, NCF2, P2RY2, PNO1, PPP4R1, PPP4R2, PTGER4, RALGAPB, RTN4, SLC35A5, SMEK1, SMEK2, SPRR2A, SRC, SRM, SRPX, TGFB1, TNFRSF11A, UBL5 3-alpha,17-beta-androstanediol, 3-beta,17-beta-androstanediol, adenosinetetraphosphatase, ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5F1, ATP5G1, ATP5H (includes EG:10476), ATP5I, ATP5J, ATP5J2, ATP5L, ATP5S, ATP6V1H, beta-estradiol, CALCA, CGA, Creb, ERF, GTF2B, H+transporting two-sector ATPase, Hat, HSD17B6, MAOA, MED21, MIR125B1, PARK2, PPP1R3F, PSENEN, RNU1-4, SYT11, ZNF Dermatological Diseases and Conditions, Genetic Disorder, Connective Tissue Disorders DNA Replication, Recombination, and Repair, Cell Cycle, Cellular Assembly and Organization Cell Cycle, Connective Tissue Development and Function, Cancer RNA Post-Transcriptional Modification, Cellular Development, Cellular Growth and Proliferation Carbohydrate Metabolism, Small Molecule Biochemistry, Cell Morphology RNA Post-Transcriptional Modification, Cellular Assembly and Organization, Cellular Compromise Post-Translational Modification, Protein Degradation, Protein Synthesis Tissue Morphology, Organismal Injury and Abnormalities, Gene Expression Infection Mechanism, Gene Expression, Cell Cycle Inflammatory Response, Cellular Movement, Connective Tissue Development and Function Energy Production, Nucleic Acid Metabolism, Small Molecule Biochemistry
23 Molecules in Network Score Focus molecules Top functions MRPL10, USP Post-Translational Modification, Dermatological Diseases and Conditions, Infectious Disease
24 Supplementary Table S7: The list of 300 differentially expressed genes after overexpression of hsa-mir-195 in MDA MB-231 cells FOL D FOL D FOL D FOL D OASL SGMS PLEKHA SNHG HIST2H2BE NOV ZNF ZNF IFIT KLF LOC SNHG TNFAIP DDIT ZNF DCP1A GKN PPP1R3C SDF2L UTX EPSTI HERC BAMBI ZCCHC RP5-1022P ZNF GADD45A HOXD IFIT LOC NRAS LOC EGR TRAF CRELD FSTL IFIH SAMD ZNF ABTB ISG CPEB NFKB C2orf ZNF KIAA TINAGL UFM EGR IGFBP MXD RNF TXNIP RHEBL CITED C9orf C7orf TMEM ZFP IRF RARRES ARL5B LOC UGCG PLCG PPP1R15A SELM SPAG IFI HERPUD NFKBIE MEIS3P ZC3HAV RELB NFKBIA ADAMTS GBP IFI ZNF ZNF AURKAPS CD TIPARP CRISPLD FLJ MAFF CCNL TUBB2A ID DNAJB EVI2B SFRS17A MT1F OTUD ZNF ZBTB PTGS CEBPD PLK TAF1D ISG C5orf USPL LOC FAM46A CCL SQSTM NCOA PMAIP SIPA1L MYLIP SPANXB DNAJC CCRN4L CBLB ZNF MAL LOXL RAB ARID5B CLIC SDC BIRC FLJ HIST1H2BE DUSP SLC30A TSC22D IL CENTA FZD VEGFC LOC PTGER KIAA ZNFX HERC ZNF JUN JUNB ZNF NFIL IER SLC25A LOC GADD45B LOC CYP1B ZNF TARDBP FAM179B
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
Prostate-targeted radiosensitization via aptamer-shrna chimeras in human. tumor xenografts
 SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
SUPPLEMENTARY DATA Prostate-targeted radiosensitization via aptamer-shrna chimeras in human tumor xenografts Xiaohua Ni 1, Yonggang Zhang 2, Judit Ribas 1, Wasim H. Chowdhury 1, Mark Castanares 1, Zhewei
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
500 New CUSABIO Antibodies at MedStore!
 BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Nature Medicine doi: /nm.2772
 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Santa Cruz Biotechnology, Inc.
 sc-24 X sc-40 X sc-41 X sc-42 X sc-44 X sc-45 X sc-46 X sc-48 X sc-50 X sc-52 X sc-59 X sc-61 X sc-70 X sc-71 X sc-73 X sc-74 X sc-109 X sc-110 X sc-111 X sc-112 X sc-113 X sc-114 X sc-122 X sc-126 X sc-131
sc-24 X sc-40 X sc-41 X sc-42 X sc-44 X sc-45 X sc-46 X sc-48 X sc-50 X sc-52 X sc-59 X sc-61 X sc-70 X sc-71 X sc-73 X sc-74 X sc-109 X sc-110 X sc-111 X sc-112 X sc-113 X sc-114 X sc-122 X sc-126 X sc-131
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Ex4-MS2. Exon 4. pcp Pre-mRNA Pre-mRNA
 A -MS2 A Exon 4 B N.E. N.E. pcp + + - + + - C N.E. N.E. pcp + + - + + - Pre-mRNA Pre-mRNA U2 U1 U4 U5 U6 U2 U1 U4 U5 U6 1 2 3 4 5 6 1 2 3 4 5 6 Supplementary Figure 1. The and complexes contain U1 and
A -MS2 A Exon 4 B N.E. N.E. pcp + + - + + - C N.E. N.E. pcp + + - + + - Pre-mRNA Pre-mRNA U2 U1 U4 U5 U6 U2 U1 U4 U5 U6 1 2 3 4 5 6 1 2 3 4 5 6 Supplementary Figure 1. The and complexes contain U1 and
GeneTex Sonderaktion: Custom Antibody Panels
 (Stand 01/2016) GeneTex Sonderaktion: Custom Antibody Panels Fünf Antikörper (in der 25 µl-größe) zum Sonderpreis von nur 420,00 (statt 545,00 ). Bitte geben Sie bei Ihrer Bestellung die Angebotsnummer
(Stand 01/2016) GeneTex Sonderaktion: Custom Antibody Panels Fünf Antikörper (in der 25 µl-größe) zum Sonderpreis von nur 420,00 (statt 545,00 ). Bitte geben Sie bei Ihrer Bestellung die Angebotsnummer
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ ΡΑΣΗ ΤΟΥ ΓΑΣΤΡΙΚΟΥ ΒΛΕΝΝΟΓΟΝΟΥ ΣΤΗ ΛΟΙΜΩΞΗ ΜΕ ΕΛΙΚΟΒΑΚΤΗΡΙ ΙΟ ΤΟΥ ΠΥΛΩΡΟΥ ΠΡΙΝ ΚΑΙ ΜΕΤΑ ΤΗ ΘΕΡΑΠΕΙΑ
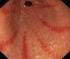 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
NCBI ID Symbol 2 A2M A4GALT A4GNT 8086 AAAS 13 AADAC AADACL AAGAB 15 AANAT 16 AARS AARS AASS 18 ABAT 19 ABCA1
 NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Supplementary Online Content
 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplementary Materials for
 www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in. and *Christina Chan
 Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
BD Cytometric Bead Array Flex Set System
 BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells
 Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma. Supplement Figure 1. Regulation of PGC1α expression in cultured podocytes.
 Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
Validità 31/01/2013 PROMO CODE
 Validità 31/01/2013 PROMO CODE SCP-P100192 SC Cat # Protein Name Application Y71-30G 14-3-3α/β Protein Western Blot Y71-30N 14-3-3α/β Protein Western Blot Y75-30G 14-3-3ε Protein Western Blot Y75-30N 14-3-3ε
Validità 31/01/2013 PROMO CODE SCP-P100192 SC Cat # Protein Name Application Y71-30G 14-3-3α/β Protein Western Blot Y71-30N 14-3-3α/β Protein Western Blot Y75-30G 14-3-3ε Protein Western Blot Y75-30N 14-3-3ε
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
 Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Burden of Disease Variants in Participants of the Long Life Family Study. Supplemental Online Material
 Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ ΩΝ ΚΑΤΑ ΤΗΝ ΕΜΒΡΥΪΚΗ ΑΝΑΠΤΥΞΗ
 ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein
 1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Κυτταρική Επικοινωνία Cell Communication Η μοίρα του κυττάρου ρυθμίζεται από εξωτερικά σήματα (από το περιβάλλον και άλλα κύτταρα) survival division differentiation apoptosis Είδη κυτταρικής επικοινωνίας
Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver
 Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Zhang et al., Supplemental figures, Methods, References and Tables
 1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
Catalog No Name Size. A Trx-tag Antibody, pab, Goat 40 μg. A GST-tag Antibody [HRP], pab, Rabbit 40 μg
![Catalog No Name Size. A Trx-tag Antibody, pab, Goat 40 μg. A GST-tag Antibody [HRP], pab, Rabbit 40 μg Catalog No Name Size. A Trx-tag Antibody, pab, Goat 40 μg. A GST-tag Antibody [HRP], pab, Rabbit 40 μg](/thumbs/83/87438311.jpg) GenScript would like to best support your research needs. For any 40ug antibody product priced at $99/vial, we re offering you the following promotion program! *Terms and Conditions This promotion is valid
GenScript would like to best support your research needs. For any 40ug antibody product priced at $99/vial, we re offering you the following promotion program! *Terms and Conditions This promotion is valid
Μοριακή βάση του καρκίνου της ουροδόχου κύστεως
 Πανεπιστήμιο Αθηνών Τμήμα Βιολογίας Τομέας Βιοχημείας & Μοριακής Βιολογίας Μοριακή βάση του καρκίνου της ουροδόχου κύστεως Μαργαρίτης Αυγέρης, Ανδρέας Σκορίλας Τομέας Βιοχημείας και Μοριακής Βιολογίας,
Πανεπιστήμιο Αθηνών Τμήμα Βιολογίας Τομέας Βιοχημείας & Μοριακής Βιολογίας Μοριακή βάση του καρκίνου της ουροδόχου κύστεως Μαργαρίτης Αυγέρης, Ανδρέας Σκορίλας Τομέας Βιοχημείας και Μοριακής Βιολογίας,
Analysis Summary: Any Allergy
 Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Debashish Sahay. To cite this version: HAL Id: tel
 Identification of genes activated and biological markers involved in lysophosphatidic acid (LPA)-induced breast cancer metastasis through its receptor LPA1 Debashish Sahay To cite this version: Debashish
Identification of genes activated and biological markers involved in lysophosphatidic acid (LPA)-induced breast cancer metastasis through its receptor LPA1 Debashish Sahay To cite this version: Debashish
Supplementary Information for. RRM2B Suppresses Activation of the Oxidative Stress Pathway and is. Up-regulated by P53 During Senescence
 Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal
 Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Nature Immunology: doi: /ni.3575
 Supplementary Figure 1 Histology of lymphoma and segregation of the mutation with the disease phenotype. (a) Histology of adenoid biopsy showing low grade B-cell lymphoma compatible with marginal zone
Supplementary Figure 1 Histology of lymphoma and segregation of the mutation with the disease phenotype. (a) Histology of adenoid biopsy showing low grade B-cell lymphoma compatible with marginal zone
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine
 Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Contents Part I Psychoneuroimmunology and Systems Biology Mechanisms 1 From Psychoneuroimmunology to Personalized, Systems, and Dynamical Medicine... 3 1.1 Psychoneuroimmunology (PNI) and Systems Biology...
Κυτταρική Επικοινωνία Cell Communication
 Κυτταρική Επικοινωνία Cell Communication Κυτταρική Επικοινωνία Είναι η ικανότητα του κυττάρου να αποκρίνεται σε εξωτερικά σήματα (σήματα από το περιβάλλον του ή άλλα κύτταρα) Τα σήματα αυτά μπορεί να είναι
Κυτταρική Επικοινωνία Cell Communication Κυτταρική Επικοινωνία Είναι η ικανότητα του κυττάρου να αποκρίνεται σε εξωτερικά σήματα (σήματα από το περιβάλλον του ή άλλα κύτταρα) Τα σήματα αυτά μπορεί να είναι
The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT
 Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supporting Information. Property focused structure-based optimization of small molecule inhibitors of the proteinprotein
 Supporting Information Property focused structure-based optimization of small molecule inhibitors of the proteinprotein interaction between menin and Mixed Lineage Leukemia (MLL) Dmitry Borkin a, Jonathan
Supporting Information Property focused structure-based optimization of small molecule inhibitors of the proteinprotein interaction between menin and Mixed Lineage Leukemia (MLL) Dmitry Borkin a, Jonathan
Supplementary Table S1 Demographic characteristics of HCC patients
 Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Information
 Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Table S2. Sequences immunoprecipitating with anti-hif-1α
 Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis
 S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
Ιωάννης Π. Τρουγκάκος Τομέας Βιολογίας Κυττάρου & Βιοφυσικής Τμήμα Βιολογίας, Παν/μιο Αθηνών ΚΥΤΤΑΡΙΚΟΣ ΚΥΚΛΟΣ - ΓΗΡΑΝΣΗ
 Ιωάννης Π. Τρουγκάκος Τομέας Βιολογίας Κυττάρου & Βιοφυσικής Τμήμα Βιολογίας, Παν/μιο Αθηνών ΚΥΤΤΑΡΙΚΟΣ ΚΥΚΛΟΣ - ΓΗΡΑΝΣΗ Σημεία ελέγχου του Κυτταρικού κύκλου (check points) ιπλασιάστηκε όλο το DNA? Είναι
Ιωάννης Π. Τρουγκάκος Τομέας Βιολογίας Κυττάρου & Βιοφυσικής Τμήμα Βιολογίας, Παν/μιο Αθηνών ΚΥΤΤΑΡΙΚΟΣ ΚΥΚΛΟΣ - ΓΗΡΑΝΣΗ Σημεία ελέγχου του Κυτταρικού κύκλου (check points) ιπλασιάστηκε όλο το DNA? Είναι
Pathway plasticity. SW2: Pathway Analysis in Transcriptomics, Proteomics and Metabolomics. Saturday, March 17, :00 am 4:30 pm.
 Pathway plasticity SW2: Pathway Analysis in Transcriptomics, Proteomics and Metabolomics Saturday, March 17, 2012 8:00 am 4:30 pm Alexander Kel Wolfenbüttel Biosoft.ru, Skolkovo Moscow alexander.kel@genexplain.com
Pathway plasticity SW2: Pathway Analysis in Transcriptomics, Proteomics and Metabolomics Saturday, March 17, 2012 8:00 am 4:30 pm Alexander Kel Wolfenbüttel Biosoft.ru, Skolkovo Moscow alexander.kel@genexplain.com
Supplementary Materials for
 www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent
 Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
The NF-κB pathways in connective tissue diseases
 κ The NF-κB pathways in connective tissue diseases TETSUO KUBOTA (Associate Professor) Tokyo Medical and Dental University Graduate School of Health Care Sciences NF-κB nuclear factor-kappa B NF-κB in
κ The NF-κB pathways in connective tissue diseases TETSUO KUBOTA (Associate Professor) Tokyo Medical and Dental University Graduate School of Health Care Sciences NF-κB nuclear factor-kappa B NF-κB in
CD4Th CD8T IL 2 NKT 1 T MHC MHC MHC. major histocompatibility complex : MHC CD28 TCR TCR. TCR αβ TCR γδ TCR. CD3 CD3 γ δ ε ζ CD4
 114 539 : TCR MHC MHC II CD4Th IFN γ Th1 IL 4 Th2 IL 17 Th17 TGFβ/IL 10 Treg IL 9 Th9 IL 22 Th22 IL 21 Tfh follicular helper MHC Ia CD8T IL 2 CD4/CD8T innate T cells MHC Ib NK TCR NKT γδ MAIMHC Ib CD8
114 539 : TCR MHC MHC II CD4Th IFN γ Th1 IL 4 Th2 IL 17 Th17 TGFβ/IL 10 Treg IL 9 Th9 IL 22 Th22 IL 21 Tfh follicular helper MHC Ia CD8T IL 2 CD4/CD8T innate T cells MHC Ib NK TCR NKT γδ MAIMHC Ib CD8
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
DrugBank. Vol. 39 Issue 13 July NECT BAI51B CXTD JYBZZ-XS-110. Tel
 * * 100102 - DrugBank - - - - 1 - - 5 2 3-4 - 2013-11-27 81373985 81173568 NECT-11-0605 2008BAI51B01 2011-6 CXTD-11 2013-JYBZZ-XS-110 * Tel 010 84738620 E-mail wangyun @ bucm. edu. cn * 010 84738620 E-mail
* * 100102 - DrugBank - - - - 1 - - 5 2 3-4 - 2013-11-27 81373985 81173568 NECT-11-0605 2008BAI51B01 2011-6 CXTD-11 2013-JYBZZ-XS-110 * Tel 010 84738620 E-mail wangyun @ bucm. edu. cn * 010 84738620 E-mail
SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in
 1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
Nature Immunology: doi: /ni Supplementary Figure 1. Foxm1 loss leads to bone marrow hypocellularity.
 Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Supplementary Figure 1 Foxm1 loss leads to bone marrow hypocellularity. (a-b) Analysis of ablation of Foxm1 as determined by semiquantitative PCR analysis of genomic DNA (a) as well as qrt-pcr analysis
Unique peptides 1 DHX9 ATP-dependent RNA Q MYBBP1A Myb-binding protein 1A Q9BQG ADAR Adenosine deaminase, Q59EC
 SUPPLEMENTAL TABLES Supplemental Table 1: List of proteins identified by mass spectrometry. # Symbol Characteristics of proteins Accession number Molecular weight (kda) Unique peptides 1 DHX9 ATP-dependent
SUPPLEMENTAL TABLES Supplemental Table 1: List of proteins identified by mass spectrometry. # Symbol Characteristics of proteins Accession number Molecular weight (kda) Unique peptides 1 DHX9 ATP-dependent
«ΧΑΡΑΚΤΗΡΙΣΜΟΣ ΜΕΤΑΓΡΑΦΙΚΩΝ ΜΟΝΟΠΑΤΙΩΝ ΠΟΥ ΣΥΝΔΕΟΝΤΑΙ ΜΕ ΤΗΝ ΑΛΛΕΡΓΙΑ (ΑΝΑΦΟΡΑ)»
 ΠΑΡΑΔΟΤΕΟ: Π.2.5 «ΧΑΡΑΚΤΗΡΙΣΜΟΣ ΜΕΤΑΓΡΑΦΙΚΩΝ ΜΟΝΟΠΑΤΙΩΝ ΠΟΥ ΣΥΝΔΕΟΝΤΑΙ ΜΕ ΤΗΝ ΑΛΛΕΡΓΙΑ (ΑΝΑΦΟΡΑ)» για το ΥΠΟΕΡΓΟ 1 της Πράξης: «ΘΑΛΗΣ- Ίδρυμα Ιατροβιολογικών Ερευνών Ακαδημίας Αθηνών- Ωρίμανση της μη ειδικής
ΠΑΡΑΔΟΤΕΟ: Π.2.5 «ΧΑΡΑΚΤΗΡΙΣΜΟΣ ΜΕΤΑΓΡΑΦΙΚΩΝ ΜΟΝΟΠΑΤΙΩΝ ΠΟΥ ΣΥΝΔΕΟΝΤΑΙ ΜΕ ΤΗΝ ΑΛΛΕΡΓΙΑ (ΑΝΑΦΟΡΑ)» για το ΥΠΟΕΡΓΟ 1 της Πράξης: «ΘΑΛΗΣ- Ίδρυμα Ιατροβιολογικών Ερευνών Ακαδημίας Αθηνών- Ωρίμανση της μη ειδικής
Supplemental Information. A p53 Super-tumor Suppressor Reveals a Tumor. Suppressive p53-ptpn14-yap Axis. in Pancreatic Cancer
 Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
ΜΔΗΕΟΝ ΤΜΠΛΔΓΜΑ ΗΣΟΤΜΒΑΣΟΣΖΣΑ
 ΜΔΗΕΟΝ ΤΜΠΛΔΓΜΑ ΗΣΟΤΜΒΑΣΟΣΖΣΑ 5 Σε όια ζρεδόλ ηα ζπνλδπισηά, ηα νπνία έρνπλ έσο ζήκεξα κειεηεζεί θαη ηδίσο ζηα ζειαζηηθά, ζπκπεξηιακβαλνκέλνπ θαη ηνπ αλζξώπνπ, έρεη δηαπηζησζεί ε ύπαξμε κηαο ζηελά ζπλδεδεκέλεο
ΜΔΗΕΟΝ ΤΜΠΛΔΓΜΑ ΗΣΟΤΜΒΑΣΟΣΖΣΑ 5 Σε όια ζρεδόλ ηα ζπνλδπισηά, ηα νπνία έρνπλ έσο ζήκεξα κειεηεζεί θαη ηδίσο ζηα ζειαζηηθά, ζπκπεξηιακβαλνκέλνπ θαη ηνπ αλζξώπνπ, έρεη δηαπηζησζεί ε ύπαξμε κηαο ζηελά ζπλδεδεκέλεο
Supplementary Table 1. Composition of the diets
 Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Κεφάλαιο 22 Γενετική του καρκίνου. Η πρωτεΐνη p53 προσδένεται στο DNA.
 Κεφάλαιο 22 Γενετική του καρκίνου Η πρωτεΐνη p53 προσδένεται στο DNA. 1 ΕΙΚΟΝΑ 22.1 Μαστογραφία που απεικονίζει έναν όγκο. Όγκος 2 ΕΙΚΟΝΑ 22.2 Ορισμένα από τα μοριακά γεγονότα που ελέγχουν τον κυτταρικό
Κεφάλαιο 22 Γενετική του καρκίνου Η πρωτεΐνη p53 προσδένεται στο DNA. 1 ΕΙΚΟΝΑ 22.1 Μαστογραφία που απεικονίζει έναν όγκο. Όγκος 2 ΕΙΚΟΝΑ 22.2 Ορισμένα από τα μοριακά γεγονότα που ελέγχουν τον κυτταρικό
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development
 Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development Isabelle Buisson, Ronan Le Bouffant, Mélinée Futel, Jean-François Riou, Muriel Umbhauer To cite this version:
Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development Isabelle Buisson, Ronan Le Bouffant, Mélinée Futel, Jean-François Riou, Muriel Umbhauer To cite this version:
Μεταφραστική έρευνα στα ΙΦΝΕ Πως η πρόοδος στην βιολογία συστημάτων οδηγεί σε νέες θεραπείες
 Μεταφραστική έρευνα στα ΙΦΝΕ Πως η πρόοδος στην βιολογία συστημάτων οδηγεί σε νέες θεραπείες Γεώργιος Κολιός MD PhD, Γαστρεντερολόγος Καθηγητής Φαρμακολογίας Δ.Π.Θ Βιολογία Συστημάτων Πρόκειται για μία
Μεταφραστική έρευνα στα ΙΦΝΕ Πως η πρόοδος στην βιολογία συστημάτων οδηγεί σε νέες θεραπείες Γεώργιος Κολιός MD PhD, Γαστρεντερολόγος Καθηγητής Φαρμακολογίας Δ.Π.Θ Βιολογία Συστημάτων Πρόκειται για μία
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Legends for Supplemental Data
 Legends for Supplemental Data Supplemental Fig. 1. Top molecular and cellular functions identified by Ingenuity Pathway Analysis after 6 h (left) and 24 h (right) of exposure of mouse pancreatic islets
Legends for Supplemental Data Supplemental Fig. 1. Top molecular and cellular functions identified by Ingenuity Pathway Analysis after 6 h (left) and 24 h (right) of exposure of mouse pancreatic islets
Differentially expressed genes of HEK293/HeLa Myst4 sirna vs. controls.
 Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Μοριακή βιολογία καρκίνου του πνεύμονα Ενότητα 1: Ογκολογία πνεύμονα. Κυριάκος Καρκούλιας, Επίκουρος Καθηγητής Σχολή Επιστημών Υγείας Τμήμα Ιατρικής
 Μοριακή βιολογία καρκίνου του πνεύμονα Ενότητα 1: Ογκολογία πνεύμονα Κυριάκος Καρκούλιας, Επίκουρος Καθηγητής Σχολή Επιστημών Υγείας Τμήμα Ιατρικής Εισαγωγή Ο καρκίνος του πνεύμονα παρουσιάζει άφθονες
Μοριακή βιολογία καρκίνου του πνεύμονα Ενότητα 1: Ογκολογία πνεύμονα Κυριάκος Καρκούλιας, Επίκουρος Καθηγητής Σχολή Επιστημών Υγείας Τμήμα Ιατρικής Εισαγωγή Ο καρκίνος του πνεύμονα παρουσιάζει άφθονες
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver.
 Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells
 Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee
Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee
