Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
|
|
|
- Δανάη Παπαϊωάννου
- 6 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals were treated with 4 intratumoral injections to the right tumor. a) Growth of distant tumors. b). Overall survival. *p<0.05. Pooled data from 2 experiments with 5 animals per group.
2 Supplementary Figure 2. Upregulation of immune-related genes in response to NDV. RNA isolated from NDV-infected tumors was subjected to gene expression analysis using the Nanostring Mouse Pancancer Immune Profiling panel. a) Type I interferon pathway genes. b) Dendritic cell function genes. c) T cell function genes. The actual genes, level of induction, and statistical significance are outlined in Supplementary Tables 1-3. Representative data from 2 experiments with 10 animals per group.
3 Supplementary Figure 3. NDV infection induces immune transcriptional signatures defining Th1 and CTL, but not Th2 responses. RNA isolated from NDV-infected tumors was subjected to gene expression analysis using the Nanostring Mouse Pancancer Immune Profiling panel. Immune cell subtypes were defined based on the transcriptional signatures developed by Bindea et al. (48). a) Heatmap showing raw abundance of immune cell types in each sample, compared to other samples. Orange indicates high abundance; blue indicates low abundance. b) Heatmap showing the correlation matrix of raw immune cell abundance. Red and blue indicate positive and negative correlation, respectively. c) Covariate plot of raw cell type abundance measurements of treated vs. control samples. Representative data from 2 experiments with 10 animals per group.
4 Supplementary Figure 4. Expression of transgene from NDV in vivo. Animals bearing flank B16-F10 tumors were injected intratumorally with NDV expressing firefly luciferase (NDV-fluc) on the indicated days and analyzed on IVIS Imaging System. a) Representative luminescence images. b) Quantified total radiance from tumor area in photons per second (p/s). c) Area under the curve (AUC) calculated from (b). Representative data from 2 experiments with 5 animals per group. Data with error bars represent mean +/- SEM. ****p<
5 Supplementary Figure 5. Expression of lineage-specific transcription factors in the CD4+FoxP3- tumor lymphocyte subsets. Top: flow cytometry plots gated on the CD4+FoxP3- (Tcon) TIL population. Bottom: Median fluorescence intensity values calculated from the plots above with average of 5 animals per group. *p<0.05, ns: non-significant. Data with error bars represent mean +/- SEM.
6 Supplementary Figure 6. Efficacy of NDV-ICOSL in non-ndv-permissive tumor cell line. MB49 tumor cells were titrated to establish a model that resulted in 100% tumor take and 0% survival in untreated animals. Animals bearing bilateral flank MB49 tumors were treated intratumorally into the right tumor with the indicated NDV and intraperitoneally with anti- CTLA-4 antibody. a) Growth of individual right (virus-injected) tumors. b) Growth of distant tumors. c) Overall survival. ns: non-significant. Pooled data from 2 experiments with 5-10 animals per group.
7 Supplementary Figure 7. Efficacy of NDV in ICOS-deficient mice. C57BL/6 ICOS-/- animals or sex and age-matched C57BL/6 controls were challenged with B16-F10 cells 1x10 5 cells in right flank and 5x10 4 cells in the left flank (a-b) or 1x10 5 cells in the left flank (c-d) and were treated intratumorally with the indicated viruses and intraperitoneal anti-ctla-4 antibody according to the schedule in figure 4b. a) Growth of individual treated and distant tumors with small tumor challenge and treatment with NDV-WT/anti-CTLA-4 combination. b) Overall survival with small tumor challenge and treatment with NDV-WT/anti-CTLA-4 combination. c) Growth of individual treated and distant tumors with larger tumor challenge and treatment with NDV-ICOSL/anti-CTLA-4 combination. d) Overall survival with small tumor challenge and treatment with NDV-ICOSL/anti-CTLA-4 combination. Pooled data from 2 experiments with 5 animals per group. ****p<
8 Supplementary Figure 8. Expression of IFNγ by tumor-infiltrating CD8+ lymphocytes in response to antigen. Splenic DC s loaded with B16-F10 or TRAMP-C2 tumor cell lysates were co-cultured with tumor T cells or naïve splenic T cells in a 1:5 (DC:lymphocyte) ratio for 8 hours. T cells were analyzed for IFNγ production by intracellular cytokine staining. Representative flow cytometry panels for each group and the controls are shown. Composite data for the analyses is shown in figure 5j.
9 Supplementary Figure 9. Composition of splenic T cell populations. Animals were treated according to the schedule in Figure 4b and spleens were collected on day 15. a) Spleen size in grams, b) Total calculated CD8+, Tcon, and Treg lymphocytes per gram of spleen. Data with error bars represent mean +/- SEM. Representative data from 2 experiments with 5 mice per group. ns: non-significant.
10 Supplementary Figure 10. Effect of NDV-ICOSL and CTLA-4 blockade on the splenic follicular helper T cells. Animals were treated according to the schedule specified in figure 4b and the spleens were collected on day 15. a) Expression of bcl-6 in CD4+FoxP3- (Tcon) subset. b) Overall percentages and gating strategy (left) and calculated absolute counts (right) of the splenic Tfh cells, as defined by CD4 + FoxP3 - PD-1 + CXCR5 + ICOS + lineage. Representative data of 2 experiments with 5 mice per group. Data with error bars represent mean +/- SEM. *p<0.05, **p<0.01,, ns: non-significant.
11 Supplementary Table 1. Upregulation of genes related to type I interferon. Gene Log2 fold change Lower confidence limit Upper confidence limit P-value Ifitm E-06 Ifih E-05 Cd3e E-05 Ifit E-05 Ciita E-05 Ifi E-05 Runx E-05 Eomes Tapbp Irf Icosl Il12rb Gzmk Sh2d1a Gbp Nfkbia Ifna Nod Irf H2.T Ifngr Tmed Irf Ubc Ifnar Ifi44l H2.Aa Ifna Ifnb Ccr Ifit Masp Cxcl Ddx Ifna Ifitm Ifi Ifi
12 Fadd Ifit
13 Supplementary Table 2. Upregulation of genes related to DC function Gene Log2 fold change Lower confidence limit Upper confidence limit P-value Ikzf E-07 Ccr E-06 Ly E-05 Cd E-05 Ccl E-05 Cxcr E-05 Cd40lg E-05 Ccr E-05 Ccl E-05 Cxcr Cr Ccr Cd Rela Cd Tfrc
14 Supplementary Table 3. Upregulation of genes related to T cell function. Gene Log2 fold change Lower confidence limit Upper confidence limit P-value Cd E-08 Ikzf E-07 Cd E-07 Tnfsf E-07 Icos E-07 Il18r E-06 Ctla E-06 Cd E-06 Btla E-06 Ifng E-06 Jak E-06 Cd E-06 Cd8a E-06 Itgal E-06 Gzmb E-06 Ccr E-06 Il E-06 Ccr E-06 Stat E-06 Itk E-06 Il12rb E-06 Cd8b E-06 Spn E-06 Il E-06 Ccl E-06 Il7r E-06 Tbx E-06 Cd E-05 Gata E-05 Ptprc E-05 Thbd E-05 Cd3e E-05 Cd E-05 Tlr E-05 Cd3d E-05 H2.K E-05 Itgam E-05 Ccl E-05 Ripk E-05
15 Cd E-05 Cd3g E-05 Dpp E-05 Cxcr E-05 Ccl E-05 Stat E-05 Tank E-05 Fut E-05 Ikzf E-05 Twist E-05 Psen E-05 Tbk E-05 Cd40lg E-05 Ccr E-05 Ccr E-05 Icam E-05 Jak E-05 Tnfrsf E-05 Il2rb E-05 Bcl E-05 Cd1d E-05 Ccl E-05 Irak E-05 Gfi Cd1d H2.D Eomes Klrk Cxcr Itch Tnfsf Nfatc Il12rb Pou2af Maf Traf Cd Stat Il6ra Cd Il
16 Gzmk Lcp Il4ra Cd Tnfrsf13b Ccl Map3k Card Tnfsf Crebbp Stat5b Rag H2.M Pdcd Socs Nod Tnfrsf1b Cd Ccl Il1rn Ncf Cd59b Tlr Snai Cma Csf Cxcl Bcl Il Tnfsf Ccr Fcgr Il6st Pax Ythdf Tlr Cd Ifnar Il Fas Nfatc
17 Irf Foxp Ido H2.Aa Fasl Relb H2.Eb Txnip Ccr Tollip Prkce Rora H2.Ab Bcl Il17rb Tnfsf13b Ctsh Map4k Il17a Cebpb Tgfb Cd Foxj Itgae Il1b Sele Il11ra Il H2.Q Tnfrsf13c Ccnd Tmem Ifna Casp Ikbkg Flt Il12b Rela Il Hamp Trem
18 St6gal Cxcl Itgb Mif Tnfsf Isg Ptgs Yy Lbp Cd Cd Vcam Usp Egr Lcn Gpr Tfrc Irf Xbp Hspb Anxa Il1a Tie Itgb Psma Pvr Tgfb Il22ra Ada Il Il12a Rorc Mapk
Supplementary Figure 1
 NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
a 2,5 b 2,5 upplemental Figure 1 IL4 (4h) in Ja18-/- mice IFN-γ (16h) in Ja18-/- mice 1,5 1,5 ng/ml ng/ml 0,5 0,5 4ClPh PyrC 4ClPh PyrC
 a 2,5 IL4 (4h) in Ja18-/- mice b 2,5 IFN-γ (16h) in Ja18-/- mice 2 2 ng/ml 1,5 ng/ml 1,5 1 1,5,5 4ClPh NC PyrC 4ClPh NC PyrC upplemental Figure 1 a b c PyrC-α-GalCer NU-α-GalCer α-galcer CD4 (PE) MFI CD4
a 2,5 IL4 (4h) in Ja18-/- mice b 2,5 IFN-γ (16h) in Ja18-/- mice 2 2 ng/ml 1,5 ng/ml 1,5 1 1,5,5 4ClPh NC PyrC 4ClPh NC PyrC upplemental Figure 1 a b c PyrC-α-GalCer NU-α-GalCer α-galcer CD4 (PE) MFI CD4
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
TABLES AND FORMULAS FOR MOORE Basic Practice of Statistics
 TABLES AND FORMULAS FOR MOORE Basic Practice of Statistics Exploring Data: Distributions Look for overall pattern (shape, center, spread) and deviations (outliers). Mean (use a calculator): x = x 1 + x
TABLES AND FORMULAS FOR MOORE Basic Practice of Statistics Exploring Data: Distributions Look for overall pattern (shape, center, spread) and deviations (outliers). Mean (use a calculator): x = x 1 + x
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks. Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks
 Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
Si + Al Mg Fe + Mn +Ni Ca rim Ca p.f.u
 .6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
.6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary Information Titles
 Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Biapenem BIPM Lot No g mg
 MexAB-OprM 1 1 1 1 16 4 9 16 5 11 BIPM imipenemcilastatinipmcs MEPM 3 MexAB-OprM BIPM IPMCS MEPM MexAB-OprM MexCD-OprJ BIPM MEPM IPMCS BIPM IPMCS MEPM 10 5 order CFUlung BIPMIPM CS MEPM 10 6 order CFUlung
MexAB-OprM 1 1 1 1 16 4 9 16 5 11 BIPM imipenemcilastatinipmcs MEPM 3 MexAB-OprM BIPM IPMCS MEPM MexAB-OprM MexCD-OprJ BIPM MEPM IPMCS BIPM IPMCS MEPM 10 5 order CFUlung BIPMIPM CS MEPM 10 6 order CFUlung
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
DC-CIK. Tca8113, [ ] (2011) CIK, Tca8113. Tca8113 DC CIK DC-CIK ; t (P=0.0132); , SAS Tca8113
![DC-CIK. Tca8113, [ ] (2011) CIK, Tca8113. Tca8113 DC CIK DC-CIK ; t (P=0.0132); , SAS Tca8113 DC-CIK. Tca8113, [ ] (2011) CIK, Tca8113. Tca8113 DC CIK DC-CIK ; t (P=0.0132); , SAS Tca8113](/thumbs/92/109031070.jpg) China Journal of Oral and Maxillofacial Surgery Vol.9 No.6 November,2011 467 [ ]1672-3244(2011)06-0467-05 DC-CIK 1 2 1 1 (1. 200011; 2. 200031) [ ] : (DC) (CIK) : DC, CIK, DC CIK DC-CIK ; Tca8113 A B C;
China Journal of Oral and Maxillofacial Surgery Vol.9 No.6 November,2011 467 [ ]1672-3244(2011)06-0467-05 DC-CIK 1 2 1 1 (1. 200011; 2. 200031) [ ] : (DC) (CIK) : DC, CIK, DC CIK DC-CIK ; Tca8113 A B C;
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation rate.3
.5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation rate.3
Smaller. 6.3 to 100 After 1 minute's application of rated voltage at 20 C, leakage current is. not more than 0.03CV or 4 (µa), whichever is greater.
 Low Impedance, For Switching Power Supplies Low impedance and high reliability withstanding 5000 hours load life at +05 C (3000 / 2000 hours for smaller case sizes as specified below). Capacitance ranges
Low Impedance, For Switching Power Supplies Low impedance and high reliability withstanding 5000 hours load life at +05 C (3000 / 2000 hours for smaller case sizes as specified below). Capacitance ranges
Aluminum Electrolytic Capacitors
 Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Figure S1. Characterization of cells and CDK4/6 inhibitors. A, FACS plot for
 Supplementary Figure legends Figure S1. Characterization of cells and CDK4/6 inhibitors. A, FACS plot for PD-1 from wildtype and PD-1-overexpressing Jurkat cells, or CD3+ CD4+ cells from PBMCs from two
Supplementary Figure legends Figure S1. Characterization of cells and CDK4/6 inhibitors. A, FACS plot for PD-1 from wildtype and PD-1-overexpressing Jurkat cells, or CD3+ CD4+ cells from PBMCs from two
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
NMBTC.COM /
 Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
Common Common Vibration Test:... Conforms to JIS C 60068-2-6, Amplitude: 1.5mm, Frequency 10 to 55 Hz, 1 hour in each of the X, Y and Z directions. Shock Test:...Conforms to JIS C 60068-2-27, Acceleration
Aluminum Electrolytic Capacitors (Large Can Type)
 Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Biostatistics for Health Sciences Review Sheet
 Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
Supplementary Table 1. Composition of the diets
 Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Μαντζούνη, Πιπερίγκου, Χατζή. ΒΙΟΣΤΑΤΙΣΤΙΚΗ Εργαστήριο 5 ο
 Κατανομές Στατιστικών Συναρτήσεων Δύο δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,...,Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ ) S σ Τ ( Χ,Y)
Κατανομές Στατιστικών Συναρτήσεων Δύο δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,...,Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ ) S σ Τ ( Χ,Y)
Μενύχτα, Πιπερίγκου, Σαββάτης. ΒΙΟΣΤΑΤΙΣΤΙΚΗ Εργαστήριο 5 ο
 Κατανομές Στατιστικών Συναρτήσεων Δύο ανεξάρτητα δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,..., Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ )
Κατανομές Στατιστικών Συναρτήσεων Δύο ανεξάρτητα δείγματα από κανονική κατανομή Έστω Χ= ( Χ, Χ,..., Χ ) τ.δ. από Ν( µ, σ ) μεγέθους n και 1 n 1 1 Y = (Y, Y,..., Y ) τ.δ. από Ν( µ, σ ) 1 n 1 Χ Y ( µ µ )
Description of the PX-HC algorithm
 A Description of the PX-HC algorithm Let N = C c= N c and write C Nc K c= i= k= as, the Gibbs sampling algorithm at iteration m for continuous outcomes: Step A: For =,, J, draw θ m in the following steps:
A Description of the PX-HC algorithm Let N = C c= N c and write C Nc K c= i= k= as, the Gibbs sampling algorithm at iteration m for continuous outcomes: Step A: For =,, J, draw θ m in the following steps:
Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών
 Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Research on Economics and Management
 36 5 2015 5 Research on Economics and Management Vol. 36 No. 5 May 2015 490 490 F323. 9 A DOI:10.13502/j.cnki.issn1000-7636.2015.05.007 1000-7636 2015 05-0052 - 10 2008 836 70% 1. 2 2010 1 2 3 2015-03
36 5 2015 5 Research on Economics and Management Vol. 36 No. 5 May 2015 490 490 F323. 9 A DOI:10.13502/j.cnki.issn1000-7636.2015.05.007 1000-7636 2015 05-0052 - 10 2008 836 70% 1. 2 2010 1 2 3 2015-03
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Χριστίνα Φεβράνογλου, Μάριος Ζωντανός, Παρασκευή Μπούρα, Σωτήρης Τσιµπούκης, Σοφία Τσαγκούλη, Ιωάννης Γκιόζος, Ανδριανή Χαρπίδου
 Χριστίνα Φεβράνογλου, Μάριος Ζωντανός, Παρασκευή Μπούρα, Σωτήρης Τσιµπούκης, Σοφία Τσαγκούλη, Ιωάννης Γκιόζος, Ανδριανή Χαρπίδου ΟΓΚΟΛΟΓΙΚΗ ΜΟΝΑ Α, Γ ΠΑΝΕΠΙΣΤΗΜΙΑΚΗ ΠΑΘΟΛΟΓΙΚΗ ΚΛΙΝΙΚΗ, ΓΝΝΘΑ ΣΩΤΗΡΙΑ ΝΕΥΡΟΕΝ
Χριστίνα Φεβράνογλου, Μάριος Ζωντανός, Παρασκευή Μπούρα, Σωτήρης Τσιµπούκης, Σοφία Τσαγκούλη, Ιωάννης Γκιόζος, Ανδριανή Χαρπίδου ΟΓΚΟΛΟΓΙΚΗ ΜΟΝΑ Α, Γ ΠΑΝΕΠΙΣΤΗΜΙΑΚΗ ΠΑΘΟΛΟΓΙΚΗ ΚΛΙΝΙΚΗ, ΓΝΝΘΑ ΣΩΤΗΡΙΑ ΝΕΥΡΟΕΝ
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Crotonols A and B, Two Rare Tigliane Diterpenoid
Statistics 104: Quantitative Methods for Economics Formula and Theorem Review
 Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Διδακτορική Διατριβή
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Διδακτορική Διατριβή Η ΕΠΙΔΡΑΣΗ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΠΑΡΕΜΒΑΣΗΣ ΥΠΟΒΟΗΘΟΥΜΕΝΗΣ ΑΠΟ ΥΠΟΛΟΓΙΣΤΗ ΣΤΗΝ ΠΟΙΟΤΗΤΑ ΖΩΗΣ ΚΑΙ ΣΤΗΝ ΑΥΤΟΦΡΟΝΤΙΔΑ ΑΣΘΕΝΩΝ ΜΕ ΚΑΡΔΙΑΚΗ
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ Διδακτορική Διατριβή Η ΕΠΙΔΡΑΣΗ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΠΑΡΕΜΒΑΣΗΣ ΥΠΟΒΟΗΘΟΥΜΕΝΗΣ ΑΠΟ ΥΠΟΛΟΓΙΣΤΗ ΣΤΗΝ ΠΟΙΟΤΗΤΑ ΖΩΗΣ ΚΑΙ ΣΤΗΝ ΑΥΤΟΦΡΟΝΤΙΔΑ ΑΣΘΕΝΩΝ ΜΕ ΚΑΡΔΙΑΚΗ
Repeated measures Επαναληπτικές μετρήσεις
 ΠΡΟΒΛΗΜΑ Στο αρχείο δεδομένων diavitis.sav καταγράφεται η ποσότητα γλυκόζης στο αίμα 10 ασθενών στην αρχή της χορήγησης μιας θεραπείας, μετά από ένα μήνα και μετά από δύο μήνες. Μελετήστε την επίδραση
ΠΡΟΒΛΗΜΑ Στο αρχείο δεδομένων diavitis.sav καταγράφεται η ποσότητα γλυκόζης στο αίμα 10 ασθενών στην αρχή της χορήγησης μιας θεραπείας, μετά από ένα μήνα και μετά από δύο μήνες. Μελετήστε την επίδραση
Μηχανική Μάθηση Hypothesis Testing
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Μηχανική Μάθηση Hypothesis Testing Γιώργος Μπορμπουδάκης Τμήμα Επιστήμης Υπολογιστών Procedure 1. Form the null (H 0 ) and alternative (H 1 ) hypothesis 2. Consider
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ Μηχανική Μάθηση Hypothesis Testing Γιώργος Μπορμπουδάκης Τμήμα Επιστήμης Υπολογιστών Procedure 1. Form the null (H 0 ) and alternative (H 1 ) hypothesis 2. Consider
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks
 98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
98 Scientific Note X : Quantitative chemical analyses of rocks with X-ray fluorescence analyzer: major and trace elements in ultrabasic rocks Kimiko Seno and Yoichi Motoyoshi,**- +, +, ;,**. -,/ Abstract:
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models
 Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
ΕΚΤΙΜΗΣΗ ΤΟΥ ΚΟΣΤΟΥΣ ΤΩΝ ΟΔΙΚΩΝ ΑΤΥΧΗΜΑΤΩΝ ΚΑΙ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΑΡΑΓΟΝΤΩΝ ΕΠΙΡΡΟΗΣ ΤΟΥ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΕΚΤΙΜΗΣΗ ΤΟΥ ΚΟΣΤΟΥΣ ΤΩΝ ΟΔΙΚΩΝ ΑΤΥΧΗΜΑΤΩΝ ΚΑΙ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΑΡΑΓΟΝΤΩΝ ΕΠΙΡΡΟΗΣ ΤΟΥ ΔΙΑΤΡΙΒΗ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ ΔΗΜΗΤΡΙΟΥ Ν. ΠΙΤΕΡΟΥ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΠΟΛΥΤΕΧΝΙΚΗ ΣΧΟΛΗ ΤΜΗΜΑ ΠΟΛΙΤΙΚΩΝ ΜΗΧΑΝΙΚΩΝ ΕΚΤΙΜΗΣΗ ΤΟΥ ΚΟΣΤΟΥΣ ΤΩΝ ΟΔΙΚΩΝ ΑΤΥΧΗΜΑΤΩΝ ΚΑΙ ΔΙΕΡΕΥΝΗΣΗ ΤΩΝ ΠΑΡΑΓΟΝΤΩΝ ΕΠΙΡΡΟΗΣ ΤΟΥ ΔΙΑΤΡΙΒΗ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ ΔΗΜΗΤΡΙΟΥ Ν. ΠΙΤΕΡΟΥ
Aquinas College. Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET
 Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΠΕΡΙΕΧΟΜΕΝΑ. Κεφάλαιο 1: Κεφάλαιο 2: Κεφάλαιο 3:
 4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Τδξάζβεζηνο θαηάιιειε γηα επεκβάζεηο ζε κνξθή πνιηνχ ή ζθφλεο; πγθξηηηθή παξνπζίαζε θπζηθψλ, ρεκηθψλ θαη κεραληθψλ ηδηνηήησλ
 Τδξάζβεζηνο θαηάιιειε γηα επεκβάζεηο ζε κνξθή πνιηνχ ή ζθφλεο; πγθξηηηθή παξνπζίαζε θπζηθψλ, ρεκηθψλ θαη κεραληθψλ ηδηνηήησλ Γεσξγία Εαραξνπνχινπ Πνιηηηθόο Μεραληθόο Γξ, Μ.Α., ΤΠΠΟΣ 1. ΠΔΡΗΛΖΦΖ Πξφζθαηεο
Τδξάζβεζηνο θαηάιιειε γηα επεκβάζεηο ζε κνξθή πνιηνχ ή ζθφλεο; πγθξηηηθή παξνπζίαζε θπζηθψλ, ρεκηθψλ θαη κεραληθψλ ηδηνηήησλ Γεσξγία Εαραξνπνχινπ Πνιηηηθόο Μεραληθόο Γξ, Μ.Α., ΤΠΠΟΣ 1. ΠΔΡΗΛΖΦΖ Πξφζθαηεο
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
519.22(07.07) 78 : ( ) /.. ; c (07.07) , , 2008
 .. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
.. ( ) 2008 519.22(07.07) 78 : ( ) /.. ;. : -, 2008. 38 c. ( ) STATISTICA.,. STATISTICA.,. 519.22(07.07),.., 2008.., 2008., 2008 2 ... 4 1...5...5 2...14...14 3...27...27 3 ,, -. " ", :,,,... STATISTICA.,,,.
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 12/06/2013 Αρ. πρωτ. 4972 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 12/06/2013 Αρ. πρωτ. 4972 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
CD4Th CD8T IL 2 NKT 1 T MHC MHC MHC. major histocompatibility complex : MHC CD28 TCR TCR. TCR αβ TCR γδ TCR. CD3 CD3 γ δ ε ζ CD4
 114 539 : TCR MHC MHC II CD4Th IFN γ Th1 IL 4 Th2 IL 17 Th17 TGFβ/IL 10 Treg IL 9 Th9 IL 22 Th22 IL 21 Tfh follicular helper MHC Ia CD8T IL 2 CD4/CD8T innate T cells MHC Ib NK TCR NKT γδ MAIMHC Ib CD8
114 539 : TCR MHC MHC II CD4Th IFN γ Th1 IL 4 Th2 IL 17 Th17 TGFβ/IL 10 Treg IL 9 Th9 IL 22 Th22 IL 21 Tfh follicular helper MHC Ia CD8T IL 2 CD4/CD8T innate T cells MHC Ib NK TCR NKT γδ MAIMHC Ib CD8
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
ΣΤΑΤΙΣΤΙΚΗ ΕΠΙΧΕΙΡΗΣΕΩΝ ΕΙΔΙΚΑ ΘΕΜΑΤΑ. Κεφάλαιο 13. Συμπεράσματα για τη σύγκριση δύο πληθυσμών
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΔΥΤΙΚΗΣ ΕΛΛΑΔΑΣ ΤΜΗΜΑ ΔΙΟΙΚΗΣΗΣ ΕΠΙΧΕΙΡΗΣΕΩΝ ΠΑΤΡΑΣ Εργαστήριο Λήψης Αποφάσεων & Επιχειρησιακού Προγραμματισμού Καθηγητής Ι. Μητρόπουλος ΣΤΑΤΙΣΤΙΚΗ ΕΠΙΧΕΙΡΗΣΕΩΝ ΕΙΔΙΚΑ ΘΕΜΑΤΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙΔΕΥΤΙΚΟ ΙΔΡΥΜΑ ΔΥΤΙΚΗΣ ΕΛΛΑΔΑΣ ΤΜΗΜΑ ΔΙΟΙΚΗΣΗΣ ΕΠΙΧΕΙΡΗΣΕΩΝ ΠΑΤΡΑΣ Εργαστήριο Λήψης Αποφάσεων & Επιχειρησιακού Προγραμματισμού Καθηγητής Ι. Μητρόπουλος ΣΤΑΤΙΣΤΙΚΗ ΕΠΙΧΕΙΡΗΣΕΩΝ ΕΙΔΙΚΑ ΘΕΜΑΤΑ
Statistics & Research methods. Athanasios Papaioannou University of Thessaly Dept. of PE & Sport Science
 Statistics & Research methods Athanasios Papaioannou University of Thessaly Dept. of PE & Sport Science 30 25 1,65 20 1,66 15 10 5 1,67 1,68 Κανονική 0 Height 1,69 Καμπύλη Κανονική Διακύμανση & Ζ-scores
Statistics & Research methods Athanasios Papaioannou University of Thessaly Dept. of PE & Sport Science 30 25 1,65 20 1,66 15 10 5 1,67 1,68 Κανονική 0 Height 1,69 Καμπύλη Κανονική Διακύμανση & Ζ-scores
ST5224: Advanced Statistical Theory II
 ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
ST5224: Advanced Statistical Theory II 2014/2015: Semester II Tutorial 7 1. Let X be a sample from a population P and consider testing hypotheses H 0 : P = P 0 versus H 1 : P = P 1, where P j is a known
Review Test 3. MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Review Test MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. Find the exact value of the expression. 1) sin - 11π 1 1) + - + - - ) sin 11π 1 ) ( -
Review Test MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. Find the exact value of the expression. 1) sin - 11π 1 1) + - + - - ) sin 11π 1 ) ( -
Supplementary Materials: Development of Amyloseand β-cyclodextrin-based Chiral Fluorescent Sensors Bearing Terthienyl Pendants
 S1 of S12 Supplementary Materials: Development of Amyloseand β-cyclodextrin-based Chiral Fluorescent Sensors Bearing Terthienyl Pendants Tomoyuki Ikai, Changsik Yun, Yutaka Kojima, Daisuke Suzuki, Katsuhiro
S1 of S12 Supplementary Materials: Development of Amyloseand β-cyclodextrin-based Chiral Fluorescent Sensors Bearing Terthienyl Pendants Tomoyuki Ikai, Changsik Yun, Yutaka Kojima, Daisuke Suzuki, Katsuhiro
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
SUPPLEMENTARY INFORMATION
 Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads. Electronic Supplementary Material
 A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads Electronic Supplementary Material Ivan Jarić 1,2*, Jörn Gessner 1 and Mirjana Lenhardt 3 1 Leibniz-Institute
A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads Electronic Supplementary Material Ivan Jarić 1,2*, Jörn Gessner 1 and Mirjana Lenhardt 3 1 Leibniz-Institute
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
 Problem 7.19 Ignoring reflection at the air soil boundary, if the amplitude of a 3-GHz incident wave is 10 V/m at the surface of a wet soil medium, at what depth will it be down to 1 mv/m? Wet soil is
Problem 7.19 Ignoring reflection at the air soil boundary, if the amplitude of a 3-GHz incident wave is 10 V/m at the surface of a wet soil medium, at what depth will it be down to 1 mv/m? Wet soil is
ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΡΓΑΣΤΗΡΙΟ ΔΙΑΧΕΙΡΙΣΗΣ ΤΟΞΙΚΩΝ ΚΑΙ ΕΠΙΚΙΝΔΥΝΩΝ ΑΠΟΒΛΗΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΡΓΑΣΤΗΡΙΟ ΔΙΑΧΕΙΡΙΣΗΣ ΤΟΞΙΚΩΝ ΚΑΙ ΕΠΙΚΙΝΔΥΝΩΝ ΑΠΟΒΛΗΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Χρήση πρότυπων τασιενεργών και οργανικών οξέων για την ηλεκτροαποκατάσταση
ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΡΓΑΣΤΗΡΙΟ ΔΙΑΧΕΙΡΙΣΗΣ ΤΟΞΙΚΩΝ ΚΑΙ ΕΠΙΚΙΝΔΥΝΩΝ ΑΠΟΒΛΗΤΩΝ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Χρήση πρότυπων τασιενεργών και οργανικών οξέων για την ηλεκτροαποκατάσταση
 1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,,, (, 100875) 1989 12 25 1990 2 23, - 2-4 ;,,, ; -
 25 3 2003 5 RESOURCES SCIENCE Vol. 25 No. 3 May 2003 ( 100875) : 500L - 2-4 - 6-8 - 10 114h - 120h 6h 1989 12 25 1990 2 23-2 - 4 : ; ; - 4 1186cm d - 1 10cm 514d ; : 714 13 317 714 119 317 : ; ; ; :P731
25 3 2003 5 RESOURCES SCIENCE Vol. 25 No. 3 May 2003 ( 100875) : 500L - 2-4 - 6-8 - 10 114h - 120h 6h 1989 12 25 1990 2 23-2 - 4 : ; ; - 4 1186cm d - 1 10cm 514d ; : 714 13 317 714 119 317 : ; ; ; :P731
Supplementary Materials for Evolutionary Multiobjective Optimization Based Multimodal Optimization: Fitness Landscape Approximation and Peak Detection
 IEEE TRANSACTIONS ON EVOLUTIONARY COMPUTATION, VOL. XX, NO. X, XXXX XXXX Supplementary Materials for Evolutionary Multiobjective Optimization Based Multimodal Optimization: Fitness Landscape Approximation
IEEE TRANSACTIONS ON EVOLUTIONARY COMPUTATION, VOL. XX, NO. X, XXXX XXXX Supplementary Materials for Evolutionary Multiobjective Optimization Based Multimodal Optimization: Fitness Landscape Approximation
DERIVATION OF MILES EQUATION FOR AN APPLIED FORCE Revision C
 DERIVATION OF MILES EQUATION FOR AN APPLIED FORCE Revision C By Tom Irvine Email: tomirvine@aol.com August 6, 8 Introduction The obective is to derive a Miles equation which gives the overall response
DERIVATION OF MILES EQUATION FOR AN APPLIED FORCE Revision C By Tom Irvine Email: tomirvine@aol.com August 6, 8 Introduction The obective is to derive a Miles equation which gives the overall response
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov.
 A. ΈΛΕΓΧΟΣ ΚΑΝΟΝΙΚΟΤΗΤΑΣ A 1. Έλεγχος κανονικότητας Kolmogorov-Smirnov. Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov. Μηδενική υπόθεση:
A. ΈΛΕΓΧΟΣ ΚΑΝΟΝΙΚΟΤΗΤΑΣ A 1. Έλεγχος κανονικότητας Kolmogorov-Smirnov. Για να ελέγξουµε αν η κατανοµή µιας µεταβλητής είναι συµβατή µε την κανονική εφαρµόζουµε το test Kolmogorov-Smirnov. Μηδενική υπόθεση:
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΜΙΚΡΟΒΙΟΛΟΓΙΚH ΠΟΙΟΤΗΤΑ ΝΩΠΟΥ ΑΓΕΛΑΔΙΝΟΥ ΓΑΛΑΚΤΟΣ: ΟΡΙΖΟΝΤΙΑ ΔΕΙΓΜΑΤΟΛΗΨΙΑ ΑΠΟ ΕΝΑ ΒΟΥΣΤΑΣΙΟ Κυριακή
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΜΙΚΡΟΒΙΟΛΟΓΙΚH ΠΟΙΟΤΗΤΑ ΝΩΠΟΥ ΑΓΕΛΑΔΙΝΟΥ ΓΑΛΑΚΤΟΣ: ΟΡΙΖΟΝΤΙΑ ΔΕΙΓΜΑΤΟΛΗΨΙΑ ΑΠΟ ΕΝΑ ΒΟΥΣΤΑΣΙΟ Κυριακή
τατιςτική ςτην Εκπαίδευςη II
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΣΙΑ ΠΑΝΕΠΙΣΗΜΙΟ ΚΡΗΣΗ τατιςτική ςτην Εκπαίδευςη II Αρχείο αποτελεςμάτων Διδάσκων: Μιχάλης Λιναρδάκης ΠΑΙΔΑΓΩΓΙΚΟ ΤΜΗΜΑ ΔΗΜΟΤΙΚΗΣ ΕΚΠΑΙΔΕΥΣΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΑΓΩΓΗΣ Άδειες Χρήσης Το παρόν
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΣΙΑ ΠΑΝΕΠΙΣΗΜΙΟ ΚΡΗΣΗ τατιςτική ςτην Εκπαίδευςη II Αρχείο αποτελεςμάτων Διδάσκων: Μιχάλης Λιναρδάκης ΠΑΙΔΑΓΩΓΙΚΟ ΤΜΗΜΑ ΔΗΜΟΤΙΚΗΣ ΕΚΠΑΙΔΕΥΣΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΑΓΩΓΗΣ Άδειες Χρήσης Το παρόν
Θωμάς ΣΑΛΟΝΙΚΙΟΣ 1, Χρήστος ΚΑΡΑΚΩΣΤΑΣ 2, Βασίλειος ΛΕΚΙΔΗΣ 2, Μίλτων ΔΗΜΟΣΘΕΝΟΥΣ 1, Τριαντάφυλλος ΜΑΚΑΡΙΟΣ 3,
 Αξιοποίηση Έξι Σεισμών στην Πελοπόννησο για την Συσχέτιση Φασματικών Επιταχύνσεων με την Απόκριση του Δομημένου Περιβάλλοντος Correlation of Spectral Accelerations with the Response of the Built Environment
Αξιοποίηση Έξι Σεισμών στην Πελοπόννησο για την Συσχέτιση Φασματικών Επιταχύνσεων με την Απόκριση του Δομημένου Περιβάλλοντος Correlation of Spectral Accelerations with the Response of the Built Environment
 n n 1 n+1 2 2 Farmers in Random Insurance Group Farmers in Random Insurance Group Insured Plots Control Plots 1st Choice Plots Insured plot Adverse Selection Moral Hazard Control plot 1st Choice Plots
n n 1 n+1 2 2 Farmers in Random Insurance Group Farmers in Random Insurance Group Insured Plots Control Plots 1st Choice Plots Insured plot Adverse Selection Moral Hazard Control plot 1st Choice Plots
Απόκριση σε Μοναδιαία Ωστική Δύναμη (Unit Impulse) Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο. Απόστολος Σ.
 Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο The time integral of a force is referred to as impulse, is determined by and is obtained from: Newton s 2 nd Law of motion states that the action
Απόκριση σε Δυνάμεις Αυθαίρετα Μεταβαλλόμενες με το Χρόνο The time integral of a force is referred to as impulse, is determined by and is obtained from: Newton s 2 nd Law of motion states that the action
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΕΠΙΔΡΑΣΗ ΥΠΕΡΗΧΩΝ ΣΤΗΝ LISTERIA GRAYI ΣΤΟ ΓΑΛΑ: ΕΠΙΒΙΩΣΗ ΚΑΙ ΦΥΣΙΚΟΧΗΜΙΚΕΣ ΕΠΙΔΡΑΣΕΙΣ. Άρτεμις
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΤΕΧΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΚΑΙ ΔΙΑΧΕΙΡΙΣΗΣ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Πτυχιακή εργασία ΕΠΙΔΡΑΣΗ ΥΠΕΡΗΧΩΝ ΣΤΗΝ LISTERIA GRAYI ΣΤΟ ΓΑΛΑ: ΕΠΙΒΙΩΣΗ ΚΑΙ ΦΥΣΙΚΟΧΗΜΙΚΕΣ ΕΠΙΔΡΑΣΕΙΣ. Άρτεμις
Tanzania. General Climate. UNDP Climate Change Country Profiles. C. McSweeney 1, M. New 1,2 and G. Lizcano 1
 UNDPClimateChangeCountryProfiles Tanzania C.McSweeney 1,M.New 1,2 andg.lizcano 1 1.SchoolofGeographyandEnvironment,UniversityofOxford. 2.TyndallCentreforClimateChangeResearch http://country-profiles.geog.ox.ac.uk
UNDPClimateChangeCountryProfiles Tanzania C.McSweeney 1,M.New 1,2 andg.lizcano 1 1.SchoolofGeographyandEnvironment,UniversityofOxford. 2.TyndallCentreforClimateChangeResearch http://country-profiles.geog.ox.ac.uk
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
PE CVVH PE+HP HP+CVVH HP+CVVH+PE CVVH. ENLAV of HCO - 3 ENLAV-HCO ± μmol /L HP+PE HP+CVVH HP+CVVH+PE
 2624 54 plasma exchange PE hemoperfusion HP continuous venovenous hemofiltration CVVH 54 PE CVVH PE+HP HP+CVVH HP+CVVH+PE 183 exceed normal limit absolute value ENLAV prothrombin time PT PE total bilirubin
2624 54 plasma exchange PE hemoperfusion HP continuous venovenous hemofiltration CVVH 54 PE CVVH PE+HP HP+CVVH HP+CVVH+PE 183 exceed normal limit absolute value ENLAV prothrombin time PT PE total bilirubin
BD Cytometric Bead Array Flex Set System
 BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ
 ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές. Εργαστήριο Γεωργίας. Viola adorata
 One-way ANOVA µε το SPSS Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές Τοµέας Φυτών Μεγάλης Καλλιέργειας και Οικολογίας, Εργαστήριο Γεωργίας Viola adorata To call in a statistician after the experiment is
One-way ANOVA µε το SPSS Επιστηµονική Επιµέλεια ρ. Γεώργιος Μενεξές Τοµέας Φυτών Μεγάλης Καλλιέργειας και Οικολογίας, Εργαστήριο Γεωργίας Viola adorata To call in a statistician after the experiment is
FORMULAS FOR STATISTICS 1
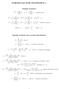 FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
