Supplementary Information
|
|
|
- Κλήμης Καψής
- 7 χρόνια πριν
- Προβολές:
Transcript
1 Supplementary Information Supplementary Figure 1: Characterization of cell surface biotinylation in non-tumorigenic and tumorigenic HCC cell lines. (a) Indicated cell lines were either mock or surface biotinylated, cell membrane fractions were enriched and affinity-purified using streptavidin-a beads and analysed by a Western blot using a streptavidin antibody. (b) Characterization of purity of cell membrane (M) and cytosolic soluble (S) fractions. Integrin β1 and Rho-GDI are markers for membrane and soluble fractions, respectively. Molecular weight markers are indicated on the side. 1
2 Supplementary Figure 2: Localization and constitutive internalization of Agrin in HCC cell lines. (a) Biochemical density gradient fractions from the indicated cell lines were analysed by Western blot for Agrin. Caveolin-1 and Flotillin-1 are markers for lipid raft membrane (LRM) and caveolae rich domains, CD-71 marks high density non-lipid raft domains of plasma membranes and Rab-5 marks the early endosomal vesicles containing fractions. (b) Membrane and soluble fractions from MIHA and Hep3B cells were isolated as described in methods section and Western blotted for Agrin. Rho-GDI and Integrin β1 served as markers for soluble and membrane fractions, respectively. (c) Hep3B cells were treated with Agrin antibody and recombinant Alexa 488 labeled cholera toxin B (CTxB) for 1h at 4 C, allowed to internalize at 37 C for the indicated time-points, fixed and processed for immunofluorescence microscopy analysis. Representative confocal images of internalization assay are shown. Boxed areas represent enlarged panels. Scale bar: 10μm. (d) Line scan signal intensity plots for the indicated cells at 4 C (cell-surface) and 37 C (internalized) are shown. Fluorescence signal intensity is represented in arbitrary units. Scale bar: 10μm. (e) Hep3B cells were incubated with Agrin antibody as in (c) and allowed to internalize for indicated time-points. 4% PFA fixed cells were immunostained with rabbit anti-eea-1 antibody for 1h at room temperature (RT), followed by anti-mouse Alexa 555 and anti-rabbit Alexa 488 secondary antibodies. Representative confocal images are shown. Boxed areas are represented as enlarged panels. Scale bar: 10μm. (f) Quantification of internalized Agrin colocalized with EEA-1 at indicated time-points measured by the number of colocalizing cells per field. Error bars represent the s.d. of means of at least five different fields containing at least 10 cells per field (**p value=0.002, students t test). 2
3 Supplementary Figure 3: Cell surface expression of Agrin in HCC cell lines. Surface biotinylated or total cell lysate from indicated cell lines were subjected to a Western blot using Agrin antibody. Actin served as loading control for total cell lysates. 3
4 Supplementary Figure 4: Anti-tumorigenic effects of sirna mediated Agrin knockdown in Hep3B cells. (a) Hep3B cells transfected with either control or Agrin sirna were cultured for the indicated number of days. At each time-point, cells were trypsinized and total cell count was measured as an index of proliferation. Error bar represents s.d. of means for three independent experiments performed in triplicates (**p value=0.002, students t test). 72 h post-transfection, total cell lysates were collected and analysed for Agrin knockdown. β-actin served as a loading control. (b) Morphology of control and Agrin sirna transfected Hep3B cells 72h post-transfection. Representative bright field microscope images are shown. Non-tumorigenic MIHA cells are used as control. Scale bar: 10 µm. (c) Control or si-agrin transfected Hep3B cells were immunostained with mouse Ki67 antibody followed by anti-mouse secondary antibody conjugated with Alexa fluor 488. Representative confocal images merged with DAPI are shown. Boxed region represents the enlarged panel with nuclear Ki67 staining. Scale bar: 10μm. Quantitative plot depicting the percentage Ki67 positive cells in at-least five different microscopic fields bearing at least cells each. Error bars represent the s.d. of the means of at least five different fields (*p value=0.03, students t test). (d) Western blot analysis for cleaved caspase-3 in control and Agrin sirna transfected Hep3B cells. β- actin served as a loading control. (e) Hep3B cells were transfected with control or Agrin sirna. 48h post-transfection, cells were subjected to a matrigel invasion assay. Invasive cells were fixed in 4% PFA after 24h, stained with 0.1% crystal violet solution and visualized in a bright-field microscope under 10X magnification. Images were quantified using ImageJ software and represented graphically. Error bar represents s.d. of the means of three biological replicates performed in triplicates (**p value=0.002, students t test). Scale bar: 50μm. 4
5 Supplementary Figure 5: Agrin clusters with invadopodia marker cortactin at invasive matrix degradation sites. (a) Control and Agrin depleted MHCC-LM3 cells were fixed, immunostained with mouse Agrin and rabbit cortactin antibodies followed by anti-mouse Alexa 555 and anti-rabbit Alexa 488 secondary antibodies. Representative confocal images are shown. Arrows point to Agrin-cortactin clusters at the cell periphery. Boxed region is represented as enlarged panels. Scale bar: 10μm. (b) Control and Agrin depleted MHCC-LM3 cells were cultured on Cy3-Gelatin for 12h, fixed and then stained with rabbit cortactin antibody followed by anti-rabbit Alexa 488 conjugated secondary antibodies. Representative confocal images are shown. Nuclei are stained with DAPI. Boxed areas are represented as enlarged panels. Arrows indicate cortactin enriched matrix degradation sites. Scale bar: 10μm. 5
6 Supplementary Figure 6: Agrin regulates mesenchymal marker signaling. (a) RT-PCR analysis of indicated genes in control and Agrin depleted MHCC-LM3 cells. GAPDH mrna levels served as endogenous control. The numbers indicate the percentage (%) reduction in expression compared to control. Transcript sizes are indicated in base pairs (bp) on the side of the gel. (b) Three hundred microgram protein from indicated cell lines were immunoprecipitated with either mouse IgG or vimentin antibody and analyzed by Western blot probed with an Agrin antibody. The blot were stripped and re-probed with vimentin antibody. Thirty microgram (10%) total cell lysate was used as input. (c) Confocal microscopy analysis of control or Agrin shrna transduced MHCC-LM3 cells with or without soluble Agrin (20μg/ml) treatment for 12h immunostained for E-cadherin and vimentin (green) by the respective mouse antibodies followed by anti-mouse Alexa 488 secondary antibodies. The cells were co-stained with DAPI (blue) and representative confocal images are shown. Scale bar: 10μm. (d) Control or Agrin sirna transfected MHCC-LM3 cells were either supplemented with or without soluble Agrin (20μg/ml) for 12h. Total cell lysates were analysed by Western blot for the indicated proteins. β-actin served as loading control. (e) Control or Agrin shrna transduced MHCC-LM3 cells were surface biotinylated and analyzed by a Western blot using vimentin antibody. Corresponding total cell lysate expression is also shown. β-actin served as loading control. (f) Hep3B cells were transfected with either control or Agrin specific sirna. 72h post transfection, total cell lysates were subjected to Western blot analysis for the indicated signaling proteins. β-actin served as loading control. 6
7 Supplementary Figure 7: Reverse co-immunoprecipitation interactions between Agrin and integrin-focal adhesion components. (a) shcontrol or shagrin MHCC-LM3 cells were immunoprecipitated with IgG or Agrin antibodies and Western blotted for integrin β1. The blot was stripped and re-probed for Agrin. 30 μg (10%) total cell lysate were used as input control. (b) shcontrol or shagrin MHCC-LM3 cells were immunoprecipitated with IgG or Agrin antibodies and Western blotted for FAK. The blot was stripped and re-probed for Agrin. 30 μg (10%) cell lysate were used as input control. Agrin depleted cell lysates were used as negative controls for IP reactions. 7
8 Supplementary Figure 8: Regulation of focal adhesion integrity by Agrin in HCC cell lines. (a) Confocal immunofluorescence analysis of focal adhesions in control and Agrin sirna transfected Hep3B cells visualized by phospho-fak (py397) rabbit antibody followed by anti-rabbit secondary antibody conjugated with Alexa fluor 488. Scale bar: 10μm. (b and c) Control or Agrin shrna MHCC-LM3 cells were immunostained with rabbit phospho-paxillin (Tyr118) (b) or rabbit vinculin (c) and mouse pfak antibodies followed by anti-rabbit Alexa fluor 555 and mouse Alexa fluor 488 antibodies. Representative confocal images are shown. Higher magnification enlarged representative view of focal adhesions are depicted in left panels. Arrows indicate colocalization between paxillin, vinculin and pfak at FAs. Scale bar: 10μm. (d) Serum starved control or Agrin knockdown MHCC- LM3 cells were trypsinized and plated on fibronectin coated plates for 2h. Post fixation, cells were stained with pfak py397. Cell elongation ratio calculated by signal intensity measurements for length and width of each cell in at least 5 different microscopic fields containing at least 10 cells each. Representative images are shown. Scale bar: 10μm. 8
9 Supplementary Figure 9: Activated FAK is localized in the vicinity of invadopodia and degraded matrix. (a) Immunofluorescence analysis performed in control and Agrin knockdown MHCC-LM3 cells 12h post scratch assay using rabbit cortactin and mouse py397fak antibodies. Representative confocal images are shown. Colocalization ratio intensity profile is also shown and boxed areas are represented as enlarged panels. Scale bar: 10μm. Mean focal adhesion-cortactin cluster length was calculated using ImageJ software and plotted graphically and the length is represented in arbitrary units. Error bars represent the s.d. of means of atleast cells per field (**p value=0.0022, students t test). (b) shcontrol or shagrin MHCC-LM3 cells were immunoprecipitated with rabbit IgG or cortactin antibodies and Western blotted for FAK. The blot was stripped and re-probed for cortactin. 30 μg (10%) total cell lysate were used as input control. * indicates the FAK specific band. (c and d) shcontrol and shagrin MHCC-LM3 cells were cultured on Cy3-Gelatin for 12h before fixation and processed for immunofluorescence using py397fak (c) or ppaxillintyr118 (d) antibodies. Nucleus is stained with DAPI. Arrows represent areas with degraded gelatin. Boxed areas are represented as enlarged panels. Arrows in enlarged panel indicate the presence of pfak/ppax within degraded gelatin matrix. Scale bar: 10μm. (e) shcontrol and shagrin MHCC-LM3 cells were cultured on Cy3-Gelatin and processed as in (c). Arrows represent activated focal adhesions (FA) at leading edges of cells in areas of matrix degradation. Scale bar: 10μm. Boxed region is represented as enlarged panels. At-least 5 different fields containing cells each were analysed to estimate the number of cells where FA associated with degraded gelatin. Error bar represents s.d. (**p value=0.0008, students t test). 9
10 Supplementary Figure 10: Agrin promotes FAK dependant invadopodia and ECM degradation. (a) Western blot analysis for FAK and Agrin knockdown in MHCC-LM3 cells. β-actin served as loading control. (b) Control sirna, FAK sirna alone or both Agrin and FAK sirna transfected MHCC-LM3 cells were plated on Cy3 gelatin for 18h before fixing and staining for phalloidin. Mean degraded areas were calculated by ImageJ software and represented in arbitrary units (*p value=0.02,**p value=0.001, students t test). Arrows indicate matrix degradation due to invadopodia. Boxed areas are represented as enlarged panels showing phalloidin and corresponding Cy3 gelatin merged images. Scale bar: 10μm. (c) Control or FAK sirna transfected MIHA cells either untreated or treated with soluble Agrin (sagrin) (10μg/ml) were grown in Cy3 gelatin for 1 day before fixation and staining with mouse pfak antibody. Boxed region is represented as enlarged panels inset. At least 5 different fields containing cells each were analysed to estimate the number of cells where FA associated with degraded gelatin. Scale bar: 10μm. Error bar represents s.d. (**p value=0.002, students t test). 10
11 Supplementary Figure 11: Impact of focal adhesion and Agrin depletion on liver cancer cell growth. (a) Western blot analysis of py397 FAK upon treatment of serum starved MHCC-LM3 cells with varying doses of PF for 1 day. β-actin served as loading control. (b) Control or Agrin depleted MHCC-LM3 cells were grown in matrigel coated chamber slides to form three dimensional spheres either in absence or presence of indicated concentrations of PF (FAK inhibitor) for 10 days. Representative bright-field images of spheres are shown (Magnification 20X). At least 5-7 microscopic fields per condition were analysed and sphere diameter was calculated using ImageJ analysis and represented in arbitrary units. The experiment was performed in triplicates and error bars represent s.d. (*p value=0.002, **p value=0.001, students t test). (c) Control and Agrin depleted MHCC-LM3 cells either untreated or treated with 0.8μM PF were subjected to a soft agar assay. Representative images showing colonies at day 10 are shown. Inset represents bright-field images of colonies. Number of colonies in at least 10 different microscopic fields were quantitated by ImageJ software and represented graphically. Experiment was performed in triplicates and error bars represent s.d. (*p value=0.03, **p value<0.002, students t test). Scale bar: 50 μm for panels b and c. 11
12 Supplementary Figure 12: Effect of Agrin depletion on actin polymerization. 20 μg protein lysates from control, Agrin depleted MHCC-LM3 cells either untreated or treated with soluble Agrin (10μg/ml) for 1 day were added to depolymerized pyrene G-actin to induce polymerization. Fluorescence was measured at 435nm for 1 hr at an interval of 1 min. Knockdown cell lysates of ArpC2/p34 subunit of Arp2/3 complex served as a positive control while lysis buffer alone served as negative control. Mean fluorescence in arbitrary units is plotted and experiments were performed in triplicates. Error bars indicate s.d. The mean fluorescence in arbitrary units at T 1/2 (~30 min) and the relative activity expressed as percentage of control lysates are shown below (*p value=0.03, **p value=0.005, ***p value=0.0003, students t test). 12
13 Supplementary Figure 13: Analysis of Agrin z transcript expression in HCC cell lines. (a) Schematic showing primers flanking Z exons where alternative splicing in neuronal Agrin is prevalent. (b) RT-PCR analysis of expression of Z-transcript of Agrin in indicated cell lines. GAPDH mrna is used as an endogenous control. M: marker lane showing transcript size in base pairs (bp). 13
14 Supplementary Figure 14: Role of Lrp4-MuSK complex in controlling Agrin related events in HCC. (a) Lrp4 and MuSK expression in a panel of HCC cell lines. Actin represents loading control. (b) MHCC-LM3 cell proliferation upon Lrp4 and MuSK knockdown using a crystal violet assay three days post-sirna treatment. Spectrophotometric absorbance was plotted. Statistical significance were calculated using a two-tailed student s t test and error bars represent the s.d. of three independent experiments performed in triplicates (**p value=0.004 and *p value=0.03, students t test). Western blot in the same transfected cells showing Lrp4 and MuSK knockdown. β-actin served as loading control. (c) Morphological analysis of MHCC-LM3 cells transfected with the indicated sirnas 3-5 days post-sirna treatment. Representative brightfield images are shown. Scale bar: 10μm. (d) Western blot analysis for Lrp4 and MuSK expression in a cohort of liver cancer patients in Singapore. GAPDH was used as loading control. *denotes tumor pairs with significant up-regulation of Lrp4 and/or MuSK in HCC tumors. 14
15 Supplementary Figure 15: Binding and function blocking properties of Agrin monoclonal antibodies. (a) Sequence alignment of human and rat Agrin C-terminal (C20) fragment illustrating the epitopes of monoclonal antibodies D2 (blue) and MAb5204 (red). Asterisks denote the conserved amino acids. (b) MHCC-LM3 cells were transfected with either vector control, full length Agrin-GFP, N-terminal Agrin GFP or C-terminal Agrin GFP constructs. 2 days post-transfection, total cell lysates were immunoprecipitated with a GFP monoclonal antibody and Western blotted using D2 or MAb5204 Agrin antibodies. GFP Western blot shows the expression of the constructs. (c) Differentiated mouse muscle C2C12 cells were pre-treated with the indicated antibodies (10μg/ml) for 12h, then either left untreated or treated with soluble Agrin (1μg/ml) for another 12h, immunoprecipitated with MuSK antibody and Western blotted using phospho-tyrosine specific 4G10 monoclonal antibody. The blot was stripped and re-probed for MuSK expression. (d) MHCC-LM3 cells treated with the indicated Agrin antibodies (10μg/ml) for 12h were immunoprecipitated with MuSK antibody and Western blotted using phospho-tyrosine specific 4G10 monoclonal antibody. The blot was stripped and re-probed for MuSK expression. 15
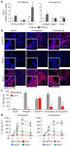
16 Supplementary Figure 16: Expression of Agrin in normal liver and HCC patient tissues. (a) HCC tissue microarray slides (US Biomax, cat #BC03116) containing normal liver and various HCC stage-wise sections were subjected to immunofluorescence analysis using anti-agrin monoclonal antibody. DNA was counter-stained with DAPI. Scale bar: 10μm. (b) Quantitative plot depicting the proportion of Agrin staining tissue positivity in different HCC stages (***p value<0.0003, students t test). The number of tumor tissues analysed for each HCC stage is indicated within parentheses. Liver cirrhosis patient tissues were used as positive control for Agrin expression. 16
17 Supplementary Figure 17: Scanned images of key Western blots used in figures 1-9. (a) Scanned uncropped blots used in figures 1-5a. (b) Scanned uncropped blots used in figures 5b-9. Boxed region represents the area cropped and molecular weight (kda) markers are indicated on the side. 17
18 Supplementary Table 1 18
19 Quantification software: MaxQuant v Search parameters: precursor tolerance - 7 ppm, MS/MS tolerance Da Modifications: Acetyl (Protein N-term), Oxidation (M), Arginine-13C615N4 (R-full), Lysine-13C615N2 (K-full) Database: HUMAN.fasta(Jan 2013) Min. Ratio Count: 2 ProPein HDs PepPide counps (Mll)ProPein nmmes Gene nmmes Sequence coqermgemol. ReigOP LkDM] Sequence lengpo RMPio HCL L3RMPio HCL normmlized L3 RMPio HCL counp L3 log2(normmlized L3) CommenPs P Apolipoprotein A-LL;Truncated apolipoprotein A-LL APOA Up Q8N5D0;Q8N5D0-2;E5RH912;2;1 Uncharacterized protein C4orf52 C4orf Up Q9HCY8 7 Protein S100-A14 S100A Up Q9NX14-2;Q9NX14 4;4 NADH dehydrogenase [ubiquinone] 1 beta subcomplex s NDUC Up Q96T88-2;Q96T88 3;3 E3 ubiquitin-protein ligase UHRC1 UHRC Up Q92804;Q ;K7EPT6;K4;4;4;1 TATA-binding protein-associated factor 2N TAC Up D3V556;D3V5Q3;P ;2;2 6.8 kda mitochondrial proteolipid C14orf2;MP Up Q96CQ6 7 Protein S100-A16 S100A Up Q96LX5 4 Up-regulated during skeletal muscle growth protein 5 USMD Up P Histone H1.5 HLST1H Up Q8ND56;Q8ND56-2;.4DTD63;3;3;2;1;1 Protein LSM14 homolog A LSM14A Up Q9Y6A9;C9J.L1 2;2 Signal peptidase complex subunit 1 SPCS Up Q9C002;H0YKW5 2;1 Normal mucosa of esophagus-specific gene 1 protein NMES1;C15orf Up Q9.UP3-3;Q9.UP3;E9PL87; 8;6;3;2 Oxidoreductase HTATLP2 HTATLP Up Q6DD88;C5H6L7;C9JUU1;C5 12;11;9;4 Atlastin-3 ATL Up P56381;Q5VTU8 3;3 ATP synthase subunit epsilon, mitochondrial;atp synthasatp5e;atp5ep Up P08670;.0YJC4;.0YJC5;Q5J 40;38;20;12;5;4;4;4 Vimentin VLM Up Q9U.L6 3 Duanine nucleotide-binding protein D(L)/D(S)/D(O) subu DND Up Q16799;Q ;Q ;1;1;1 Reticulon-1 RTN Up P82979;H0YHD0;C8VZQ9;Q56;5;5;5;1 SAP domain-containing ribonucleoprotein SARNP;CLP Up Q6P S ribosomal protein L54, mitochondrial MRPL Up Q14004;Q ;4 Cyclin-dependent kinase 13 CDK Up Q10472;C5DY99;K7EJV8 10;10;3 Polypeptide N-acetylgalactosaminyltransferase 1;PolypeDALNT Up Q96SL4 2 Dlutathione peroxidase 7 DPX Up Q9.W72 2 HLD1 domain family member 2A HLDD2A Up Q14247;Q ;Q ;16;15;7;6;4;3;3;3 Src substrate cortactin CTTN Up Q Transmembrane 9 superfamily member 2 TM9SC Up Q9Y6H1;Q5T1J5 4;3 Coiled-coil-helix-coiled-coil-helix domain-containing prochchd2;chchd2p Up Q9.TD8;K7EP90;Q9.TD8-3;2;2;2;2;2;2;1 RNA-binding protein 42 R.M Up P00533;E9PCD7;Q504U8;P038;37;34;20;20;14;5;1Epidermal growth factor receptor EDCR Up Q9P2M7;Q9P2M7-2;A6PVU5;2;1 Cingulin CDN Up O00400;H7C562 4;2 Acetyl-coenzyme A transporter 1 SLC33A Up Q9NZW5;.8ZZD1;C9J4Q3;C7;7;4;2 MADUK p55 subfamily member 6 MPP Up P33947;P ;H7.YC7 5;4;1 ER lumen protein retaining receptor 2 KDELR Up P MARCKS-related protein MARCKSL Up Q8LV08 6 Phospholipase D3 PLD Up Q9H0U4;E9PLD0;Q ;8;7 Ras-related protein Rab-1.;Putative Ras-related protein RA.1.;RA.1C Up P ;Q5VU66;Q5VU59 24;22;22;20;20;18;18;12;10;6;6;5;4;4 TPM Up Q6UW68;K7ELQ9;K7EPR0;K3;3;2;2;1;1 Transmembrane protein 205 TMEM Up Q99442;C8WC48;C8WCJ7;C54;4;3;1 Translocation protein SEC62 SEC Up Q7Z3K3;.7Z.Y5;Q7Z3K3-3;Q9;9;9;9;9;9;6;2;2;1;1;1Pogo transposable element with ZNC domain PODZ Up P Ras-related protein Rab-3. RA Up Q9.XK5;.7Z238;Q9.XK5-2 1;1;1.cl-2-like protein 13.CL2L Up D6RE79;Q14728;D6RA47;D62;2;2;2 Major facilitator superfamily domain-containing protein MCSD Up Q9U.R2 3 Cathepsin Z CTSZ Up P60602;P ;1 Reactive oxygen species modulator 1 ROMO Up Q9NRK6 7 ATP-binding cassette sub-family. member 10, mitochon A.C Up P ;P07355;H0YN42;H17;17;15;14;14;12;12;Annexin A2;Annexin;Putative annexin A2-like protein ANXA2;ANXA2P Up Q Lnositol 1,4,5-trisphosphate receptor type 3 LTPR Up P35251;P ;H0Y8U4;E5;5;1;1 Replication factor C subunit 1 RCC Up P50552;K7EM16;K7EQD0 4;2;1 Vasodilator-stimulated phosphoprotein VASP Up Q9NRZ7;Q9NRZ7-3;Q9NRZ75;4;4;4;1;1;1 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma ADPAT Up Q12955;Q ;Q ;24;24;12;5;4;4;4;4 Ankyrin-3 ANK Up O ;O ;2 ER lumen protein retaining receptor 3 KDELR Up O ;O95197;O ;4;4;4;4;3;1;1;1 Reticulon-3 RTN Up Q12849;H0YAK1;H0Y8R1;C53;3;3;3;2;1 D-rich sequence factor 1 DRSC Up Q15293;.7Z1M1;E9PJD5;E911;10;4;3;3 Reticulocalbin-1 RCN Up Q14011;D6W5Y5;.4E2X2;K75;4;4;4;4;4;4;4;4;4;4;1Cold-inducible RNA-binding protein CLR.P Up J3KQ66;P78509;P ;P 44;44;44;44 Reelin RELN Up O96000;H3.PJ9;Q96LL6;H3.5;5;4;3 NADH dehydrogenase [ubiquinone] 1 beta subcomplex s NDUC Up Q92793;Q ;2 CRE.-binding protein;histone acetyltransferase p300 CRE..P;EP Up Q7Z7H5;Q7Z7H5-2;Q7Z7H5 6;6;5;3 Transmembrane emp24 domain-containing protein 4 TMED Up O95563;Q5R3.4 7;7.rain protein 44.RP Up Q Mitochondrial import receptor subunit TOM20 homolog TOMM Up Q9H089;H7C2X7;C8WCC6 3;2;1 Large subunit DTPase 1 homolog LSD Up A6NMH8;E9PLC1;P60033;H04;4;4;4;4;4;4;1;1 CD81 antigen CD Up Q6PKD0;Q6PKD0-3;E5RH507;7;4;2;2;2;2;2;2;2;2;1La-related protein 1 LARP Up O43504;E9PLX3 4;3 Hepatitis. virus X-interacting protein H.XLP Up O15397;O ;C5H2L3 3;2;1 Lmportin-8 LPO Up Q9P2X0-2;Q9P2X0 2;2 Dolichol-phosphate mannosyltransferase subunit 3 DPM Up O14832;.1ALH6;A8MTS8;C95;5;3;2;2 Phytanoyl-CoA dioxygenase, peroxisomal PHYH Up P78332;E9PDM9;C8WCA5;.3;3;3;3 RNA-binding protein 6 R.M Up Q14697;C5H6X6 53;47 Neutral alpha-glucosidase A. DANA Up P HLA class L histocompatibility antigen, A-68 alpha chain HLA-A Up P61165;C5DWH5 1;1 UPC0197 transmembrane protein C11orf10 C11orf Up Q Spectrin alpha chain, brain SPTAN Up P55011;P ;D3XAL9;E33;32;29;1;1;1;1;1 Solute carrier family 12 member 2 SLC12A Up L3L0Q9;P04062;P ;J37;7;7;7;6;6;4 Dlucosylceramidase D.A Up P39060;P ;P ;27;27;17;12 Collagen alpha-1(xvlll) chain;endostatin COL18A Up Q8NCC6 15.iorientation of chromosomes in cell division protein 1-l.OD1L Up P27487;C8WE17;C8W..6;H 25;5;3;2 Dipeptidyl peptidase 4;Dipeptidyl peptidase 4 membranedpp Up E7EX17;P23588;.4DS13;C8V4;4;4;2;1;1;1 Eukaryotic translation initiation factor 4. ELC Up O00468;H0Y5U1 16;2 Agrin ADRN Up Q6L8Q7;Q6L8Q7-2;C6T1Q0 4;4;4 2,5-phosphodiesterase 12 PDE Up Q99856;A6NKC2 3;1 AT-rich interactive domain-containing protein 3A ARLD3A Up Q5VV89;O14880;Q5VV87 4;4;2 Microsomal glutathione S-transferase 3 MDST Up Q9NX.9;C6SH78;Q9NYP7;E 3;1;1;1 Elongation of very long chain fatty acids protein 2 ELOVL Up Q9.YC9;.7Z746;L3L2X1 7;2;2 39S ribosomal protein L20, mitochondrial MRPL Up Q7Z6E9;Q7Z6E9-2;H3.SK8;Q3;3;1;1 E3 ubiquitin-protein ligase R..P6 R..P Up Q5SSJ5;Q5SSJ5-2;Q5SWC8; 4;3;3;3;2;1;1;1 Heterochromatin protein 1-binding protein 3 HP1.P Up Q9HA82;H0YN04;H0YNR6;H3;3;3;3;3;2;2 Ceramide synthase 4 CERS Up P Nestin NES Up C9JC84;P02679;C9JEU5;P025;5;5;5;2;2 Cibrinogen gamma chain CDD Up Q9HD45;Q5T.53 7;6 Transmembrane 9 superfamily member 3 TM9SC3;RP11-34E Up O00116;.8ZZ81 20;5 Alkyldihydroxyacetonephosphate synthase, peroxisoma ADPS Up A8MV58;Q16643;Q ;20;17;15;7;5;5;3 Drebrin D.N Up Q13445;K7EQ63;K7ELN4 4;2;2 Transmembrane emp24 domain-containing protein 1 TMED Up P ;P ;P ;14;14;14;14;14;14;Protein SON SON Up P20674;H3.NX8;H3.V69;H37;6;5;4;1 Cytochrome c oxidase subunit 5A, mitochondrial COX5A Up Q13123;E7EQZ7;D6REL4 4;1;1 Protein Red LK Up O43676;C9JKQ2 2;1 NADH dehydrogenase [ubiquinone] 1 beta subcomplex s NDUC Up 19
20 F2Z2X0;Q4TT34;O00746;H0 3;3;3;2;2;1;1 Nucleoside diphosphate kinase;nucleoside diphosphate NME Up F8VX04;Q9H2H9 2;2 Sodium-coupled neutral amino acid transporter 1 SLC38A Up O14776;O ;G3V220 12;12;9 Transcription elongation regulator 1 TCERG Up Q9UGP8 3 Translocation protein SEC63 homolog SEC Up Q9BQ S ribosomal protein L34, mitochondrial MRPL Up Q8N8S7;Q8N8S7-2;Q9UI08 10;10;1 Protein enabled homolog ENAH Up F5H301;Q9UDY2;Q9UDY2-330;30;30;29;26;26;24;Tight junction protein ZO-2 TJP Up P ;P45880;P ;6;6;5;3;3;1 Voltage-dependent anion-selective channel protein 2 VDAC Up P20336;Q96E17 4;1 Ras-related protein Rab-3A RAB3A Up Q27J81;Q27J81-2;H7BXE4;Q45;44;17;7 Inverted formin-2 INF Up Q92930;H0YNE9;F5GY21;H08;7;6;4;2;2 Ras-related protein Rab-8B RAB8B Up Q9UKR5 4 Probable ergosterol biosynthetic protein 28 C14orf Up Q9Y520-7;Q9Y520;Q9Y520-54;4;4;4;4;4;4;4 Protein PRRC2C PRRC2C Up Q9H7F0;Q9H7F0-2 9;6 Probable cation-transporting ATPase 13A3 ATP13A Up O14967;D6RAZ4 9;4 Calmegin CLGN Up Q9P0S9 3 Transmembrane protein 14C TMEM14C Up Q02978;I3L1P8;F5GY65 10;10;8 Mitochondrial 2-oxoglutarate/malate carrier protein SLC25A Up Q9BU76;Q9BU76-4;Q9BU762;2;2;1 Multiple myeloma tumor-associated protein 2 MMTAG Up Q5T7F6;Q9P0B6 2;2 Coiled-coil domain-containing protein 167 CCDC Up P Protein FAM165B FAM165B Up O14521;E9PIC0;E9PIG3;H3B3;3;1;1;1;1;1;1 Succinate dehydrogenase [ubiquinone] cytochrome b smsdhd Up Q8WWM7;Q8WWM7-3;H3B7;6;6;6;6;6;6;6;6;3;3;2Ataxin-2-like protein ATXN2L Up Q13136;Q ;E9PJZ7;E4;4;3;2;1;1;1;1;1;1;1;1Liprin-alpha-1 PPFIA Up Q09666;Q ;E9PLK4;E203;3;3;3;3;3;3;3;1 Neuroblast differentiation-associated protein AHNAK AHNAK Up F5H1Z9;Q ;E9PGZ1;Q5;5;5;5;4;4;4;4;4;4 Caldesmon CALD Up O75947;O ;F5H608 13;9;4 ATP synthase subunit d, mitochondrial ATP5H Up Q96EH3 3 Mitochondrial assembly of ribosomal large subunit protemalsu Up P63173;J3QL01;J3KT73;J3KS6;5;5;3 60S ribosomal protein L38 RPL Up E9PDI4;O00515;H0Y901 2;2;1 Ladinin-1 LAD Up O S ribosomal protein L33, mitochondrial MRPL Up Q9H8G2;Q9H8G2-2 1;1 Caspase activity and apoptosis inhibitor 1 CAAP Up Q99643;Q ;Q ;4;4 Succinate dehydrogenase cytochrome b560 subunit, mitosdhc Up P35221;P ;G3XAM7; 51;49;45;44;30;13;11;Catenin alpha-1 CTNNA Up P ;P35520;F5H2U1;H6;6;5;4;2;1;1 Cystathionine beta-synthase CBS Up Q32M63;Q6ZMG9 2;2 Ceramide synthase 6 LASS6;CERS Up D6RF35;P ;P02774;P17;16;16;14;9;2 Vitamin D-binding protein GC Up E7EW20;Q9UM54-6;Q9UM58;8;7;7;7;7;7;1 Unconventional myosin-vi MYO Up Q9BYC8;B3KQX1 6;1 39S ribosomal protein L32, mitochondrial MRPL Up Q9BUN8;Q9BUN8-2;B4E1G15;5;5;2 Derlin-1 DERL Up Q9BTC0 3 Death-inducer obliterator 1 DIDO Up Q Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 LPGAT Up Q5VW38;G5E994;Q5VW38-4;4;4;2 Protein GPR107 GPR Up P ATP synthase protein 8 MT-ATP Up Q9Y3R5;Q9Y3R5-2 2;2 Protein dopey-2 DOPEY Up Q9BW60;B4DP24 3;3 Elongation of very long chain fatty acids protein 1 ELOVL Up O95202;O ;O ;6;6 LETM1 and EF-hand domain-containing protein 1, mitocholetm Up P DNA-directed RNA polymerases I, II, and III subunit RPABPOLR2L Up P67936;P ;K7ENT6;K18;15;11;10;6;5;4 Tropomyosin alpha-4 chain TPM Up P13073;H3BPG0;H3BNV9;H 8;6;5;5;4;1 Cytochrome c oxidase subunit 4 isoform 1, mitochondrialcox4i Up E1P660;J3QRS9;O43670;O4 3;3;3;3;2;2;1;1;1;1;1 Zinc finger protein 207 ZNF Up P21589;P ;Q96B60;H8;8;5;1;1 5-nucleotidase NT5E Up P39748;F5H1Y3;I3L3E9 2;1;1 Flap endonuclease 1 FEN Up Q8IZE3;Q8IZE3-2 4;4 Protein-associating with the carboxyl-terminal domain o SCYL Up Q9NS69 3 Mitochondrial import receptor subunit TOM22 homolog TOMM Up P28715;P ;1 DNA repair protein complementing XP-G cells ERCC Up P21397;B4DF46 4;3 Amine oxidase [flavin-containing] A MAOA Up Q4U2R6 3 39S ribosomal protein L51, mitochondrial MRPL Up Q9HB90;Q9NQL2;Q9NQL2-4;3;2 Ras-related GTP-binding protein C;Ras-related GTP-bindi RRAGC;RRAGD Up C9JLE3;P51654;G3V1R0;H0Y9;9;7;6;1 Glypican-3;Secreted glypican-3 GPC Up O ;O00214;F6V2D4;E2;2;2;1;1;1;1;1;1 Galectin-8 LGALS Up Q96G23;Q5SZE1;H0YNU7;Q8;7;6;5;5;5;4;3;3 Ceramide synthase 2 CERS2;LASS Up Q96Q06-2;Q96Q06 10;10 Perilipin-4 PLIN Up Q ;Q14157;F8W726; 7;7;7;7;7;7;4;2;2;2;2;1Ubiquitin-associated protein 2-like UBAP2L Up P12270;Q5SWX9 119;5 Nucleoprotein TPR TPR Up Q01740;B7Z3P4;Q5QPT3 4;2;2 Dimethylaniline monooxygenase [N-oxide-forming] 1 FMO Up O14976;E9PGR2;H0Y8H5;D63;3;1;1;1;1;1 Cyclin-G-associated kinase GAK Up Q9UQ35;Q9UQ35-2;I3L2L6; 35;19;19;7;5;5;5;4;1 Serine/arginine repetitive matrix protein 2 SRRM Up Q14315;Q ;29 Filamin-C FLNC Up Q96GC5;B4DN34;F5H702;F56;6;6;2 39S ribosomal protein L48, mitochondrial MRPL Up Q9UBT7;Q9UBT7-2;Q9UBT74;4;3;1;1 Alpha-catulin CTNNAL Up Q Endonuclease G, mitochondrial ENDOG Up P08648;H0YIV7 18;3 Integrin alpha-5;integrin alpha-5 heavy chain;integrin alpitga Up D3DNT2;E7EUU4;E9PGM1;E41;41;41;41;40;40;40;Eukaryotic translation initiation factor 4 gamma 1 EIF4G Up P16383;P ;2 GC-rich sequence DNA-binding factor 2 GCFC Up Q9UHQ9;H7C0R7 10;9 NADH-cytochrome b5 reductase 1 CYB5R Up P ;P42771;J3QRG6;K3;3;3;2;2;2;1;1;1 Cyclin-dependent kinase inhibitor 2A, isoforms 1/2/3 CDKN2A Up P Cytochrome c-type heme lyase HCCS Up Q01628;E9PS44;Q01629;P133;3;2;2;2;2 Interferon-induced transmembrane protein 3;Interferon IFITM3;IFITM2;IFIT Up O15162;C9J7K9;B4DTE8;C9J2;2;2;1;1;1;1 Phospholipid scramblase 1 PLSCR Up Q5HYI8;C9JXM3;F8WDC7;F 2;1;1;1;1;1 Rab-like protein 3 RABL Up O75964;E9PN17 6;6 ATP synthase subunit g, mitochondrial ATP5L Up Q13084;Q4TT38;A2IDC6;Q411;11;10;9;7 39S ribosomal protein L28, mitochondrial MRPL Up A4D1E9;A4D1E9-2;C9JEQ8;C6;5;4;4;4;2;2;2;2 GTP-binding protein 10 GTPBP Up P ATP synthase subunit e, mitochondrial ATP5I Up Q9Y320;Q9Y320-2;G3V155;E9;8;4;4;4;4;4 Thioredoxin-related transmembrane protein 2 TMX Up Q96DA6;C9JBV1;F2Z3A7;G57;5;4;3 Mitochondrial import inner membrane translocase subundnajc Up P24539;Q5QNZ2 16;14 ATP synthase subunit b, mitochondrial ATP5F Up G3V4K0;O95406;G3V3B2;G31;1;1;1 Protein cornichon homolog CNIH Up Q9Y666;H0YB78;Q9Y666-2;Q15;6;3;1;1 Solute carrier family 12 member 7 SLC12A Up G3V180;G3V1D3;Q9NY33;Q2;2;2;2;1;1;1;1 Dipeptidyl peptidase 3 DPP Up Q8TAT6-2;Q8TAT6;B4DG89 2;2;1;1;1 Nuclear protein localization protein 4 homolog NPLOC Up O00192;E9PDC3;O ;C2;2;2;2 Armadillo repeat protein deleted in velo-cardio-facial sy ARVCF Up O75376;O ;E7EVK1;B4;3;2;2;2;2;2;1 Nuclear receptor corepressor 1 NCOR Up Q96A33;Q96A33-2;J3QR89 14;13;3 Coiled-coil domain-containing protein 47 CCDC Up O75352;J3QRD5;J3QW43;B44;4;4;4;4;4;4;3;3;3;2;2Mannose-P-dolichol utilization defect 1 protein MPDU Up O75911;O ;Q5SUY4 10;4;4 Short-chain dehydrogenase/reductase 3 DHRS Up P49768;P ;E7ES96;P44;4;4;3;2;2;2;2;2;1;1;1Presenilin-1;Presenilin-1 NTF subunit;presenilin-1 CTF supsen1;psen Up Q86Y39;C9JT23;Q86Y39-2;K5;3;3;3;3;3;3 NADH dehydrogenase [ubiquinone] 1 alpha subcomplex NDUFA Up Q9NRY2;Q9NRY2-2 3;1 SOSS complex subunit C SSBIP Up E9PDL9;B5M453;Q9Y6M7;E 2;2;2;2;2;2;2;2;2;2;2;2Sodium bicarbonate cotransporter 3 SLC4A Up Q8TCC3-2;Q8TCC3;Q8TCC3 3;3;2;2 39S ribosomal protein L30, mitochondrial MRPL Up Q8NBM4;Q8NBM4-2;Q8NB5;4;4;4;1;1 Ubiquitin-associated domain-containing protein 2 UBAC Up O43490;O ;O ;8;8;8;8;8;8;1;1 Prominin-1 PROM Up O95865;Q5SRR8;Q5SSV3;A34;3;3;3;2 N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 DDAH Up P07686;Q5URX0;H0Y9B6;H05;4;4;3 Beta-hexosaminidase subunit beta;beta-hexosaminidasehexb Up Q10471;G3V1S6;B7Z8V8 7;7;5 Polypeptide N-acetylgalactosaminyltransferase 2;PolypeGALNT Up Q96RT1-8;Q96RT1;Q96RT1-4;4;4;4;4;4;4;4;4;3 Protein LAP2 ERBB2IP Up Q9HD33;Q9HD33-2;Q9HD339;9;5 39S ribosomal protein L47, mitochondrial MRPL Up G8JLI3;Q5H9R7-5;Q5H9R7;Q4;4;4;4;4;4;4;4;4;3;2;2Serine/threonine-protein phosphatase 6 regulatory subuppp6r Up Q4G0N4;Q4G0N4-2;B7Z8V714;12;10;10;3;2 NAD kinase domain-containing protein 1 NADKD Up E9PR17;P13987;E9PNW4;H04;4;4;2 CD59 glycoprotein CD Up 20
21 A6NH11 3 Glycolipid transfer protein domain-containing protein 2 GLTPD Up Q9BQC6 1 Ribosomal protein 63, mitochondrial MRP Up Q9NPL8;G3XA94;C9JU35;C96;3;3;2;1;1;1;1 Translocase of inner mitochondrial membrane domain-cotimmdc Up Q969N2;Q969N2-5;G8JLF5; 6;6;4;4;4;3;3;2 GPI transamidase component PIG-T PIGT Up Q ;Q14160;Q ;24;21;7 Protein scribble homolog SCRIB Up Q14108;E7EM68 2;1 Lysosome membrane protein 2 SCARB Up O15047;C9J2Z9 4;3 Histone-lysine N-methyltransferase SETD1A SETD1A Up Q92797;Q ;14 Symplekin SYMPK Up O00461;F8W785 6;6 Golgi integral membrane protein 4 GOLIM Up Q5BJD5;E9PJ42;Q5BJD5-2;E6;4;4;4 Transmembrane protein 41B TMEM41B Up P ;P ;P ;5;5;5;5;5;3;3;1;1 Clusterin;Clusterin beta chain;clusterin alpha chain;clustclu Up Q9BTT6;Q9BTT6-2;Q5T0G3 11;5;4 Leucine-rich repeat-containing protein 1 LRRC Up Q9P032 6 NADH dehydrogenase [ubiquinone] 1 alpha subcomplex NDUFAF Up O75616;J3QRV9;J3QSB2;O75;4;4;4;2;1 GTPase Era, mitochondrial ERAL Up H0YK83;P67812;H0YK72;H0 5;5;5;5;3;3;3;2 Signal peptidase complex catalytic subunit SEC11A SEC11A;SEC11L Up Q13393;Q ;Q ;5;5;3;2;1 Phospholipase D1 PLD Up Q9H9B4;D6RFI0;D6RDG7;D69;5;4;2 Sideroflexin-1 SFXN Up Q16718;Q5H9R2;H7BYD0;C96;6;6;3;3 NADH dehydrogenase [ubiquinone] 1 alpha subcomplex NDUFA5;DKFZp Up P ;J3KN01;A8MQ02;42;42;42;42;42;42;39;Afadin MLLT Up Q Sodium-coupled neutral amino acid transporter 3 SLC38A Up Q96A26;F8W7Q4;E9PH05 7;7;5 Protein FAM162A FAM162A Up Q8NHG7 2 Small VCP/p97-interacting protein SVIP Up E2QRJ6;Q9H1A6;P52435;Q93;3;3;3;2;2;2;1;1;1;1;1DNA-directed RNA polymerase II subunit RPB11-a;DNA-dPOLR2J3;POLR2J Up Q6PD62;H0YCE8 14;4 RNA polymerase-associated protein CTR9 homolog CTR Up Q96KR1;H0Y8W1 3;1 Zinc finger RNA-binding protein ZFR Up P55285;P ;D6RF86 5;4;4 Cadherin-6 CDH Up J3KQ83;P18859;A8MUH2 4;4;2 ATP synthase-coupling factor 6, mitochondrial ATP5J Up Q96JM3 3 Chromosome alignment-maintaining phosphoprotein 1 CHAMP Up P36542;P ;B4DL14 9;8;8 ATP synthase subunit gamma, mitochondrial;atp synthasatp5c Up E7ERS3;Q86VM9;Q86VM9-24;4;4;1;1 Zinc finger CCCH domain-containing protein 18 ZC3H Up Q9UHA4;J3KMX4 4;4 Ragulator complex protein LAMTOR3 LAMTOR Up P49790;F6QR24 23;22 Nuclear pore complex protein Nup153 NUP Up P51809;P ;P ;4;4;1 Vesicle-associated membrane protein 7 VAMP Q8WVK2;B8ZZ98 3;1 U4/U6.U5 small nuclear ribonucleoprotein 27 kda proteinsnrnp P35610;A8K3P4;B4DU95;B15;4;4;3;1 Sterol O-acyltransferase 1 SOAT Q9NYK5-2;Q9NYK5;C9JG87 9;9;8 39S ribosomal protein L39, mitochondrial MRPL F5H2M7;Q9H4A3-6;Q9H4A314;14;14;14;14;14;11;Serine/threonine-protein kinase WNK1 WNK Q96S52;Q96S52-2;F5H019 6;6;4 GPI transamidase component PIG-S PIGS P11233;H7C3P7;C9JPE8 6;4;2 Ras-related protein Ral-A RALA P42224;J3KPM9;P ;D3;3;3;1;1;1 Signal transducer and activator of transcription 1-alpha/bSTAT Q8TBQ9 2 Protein kish-a TMEM167A Q9Y2R0;K7EPV0 7;6 Coiled-coil domain-containing protein 56 CCDC O60437;K7EKI8;K7EQ71;REV49;49;30;1;1;1;1;1;1 Periplakin PPL P Myristoylated alanine-rich C-kinase substrate MARCKS Q16795;H3BRM9;F5H0J3 8;4;2 NADH dehydrogenase [ubiquinone] 1 alpha subcomplex NDUFA Q9Y6C9;E9PIE4;E9PNT8;F5H11;8;7;3 Mitochondrial carrier homolog 2 MTCH REV Q9C000;REV Q9C002;2;2;2;2; O Sphingolipid delta(4)-desaturase DES1 DEGS Q9Y3B7;Q9Y3B7-2;A6NLT0; 11;9;7;3;2 39S ribosomal protein L11, mitochondrial MRPL B8ZZS0;Q9NYM9;H7BXT7;E 3;3;3;2;1 BET1-like protein BET1L P50148;B1AM21 9;3 Guanine nucleotide-binding protein G(q) subunit alpha GNAQ P16615;P ;P ;37;37;37;35;32;13;Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 ATP2A P48047;H7C0C1;H7C068;H712;5;3;3 ATP synthase subunit O, mitochondrial ATP5O J3KQW8;A8MYQ9;Q9BZG1; 11;11;11;10;10;10;9;8Ras-related protein Rab-34 RAB O14828;O ;4 Secretory carrier-associated membrane protein 3 SCAMP P Cytochrome c1, heme protein, mitochondrial CYC Q13330;E7ESY4;Q ;H5;5;5;5;2;2;2;1;1;1 Metastasis-associated protein MTA1 MTA Q7Z3C6;Q7Z3C6-2;H7C1G6;4;4;4;2 Autophagy-related protein 9A ATG9A P05067;P ;P ;7;7;7;7;7;6;5;5;5;5;5Amyloid beta A4 protein;n-app;soluble APP-alpha;SolubAPP Q9BR76;E7EW44;F5H390;F59;6;6;4;1;1 Coronin-1B CORO1B P78537;P ;G8JLQ3;F 3;3;3;1;1;1;1 Biogenesis of lysosome-related organelles complex 1 subbloc1s Q9HC07;B4DHW1;D6RD79; 3;2;1;1 Transmembrane protein 165 TMEM H7C2I1;Q99873;G5E9B6;Q9 19;19;19;19;19;19;12;Protein arginine N-methyltransferase 1 PRMT P Signal peptidase complex subunit 3 SPCS Q9NVI7;Q9NVI7-2;H0Y2W28;8;7;7;4;4;4;3;2;1 ATPase family AAA domain-containing protein 3A;ATPaseATAD3A;ATAD3B; Q8IXM3 7 39S ribosomal protein L41, mitochondrial MRPL P Guanine nucleotide-binding protein G(I)/G(S)/G(O) subu GNG Q9UHB6-4;Q9UHB6;Q9UHB7;7;4;4;4;4;4;1;1 LIM domain and actin-binding protein 1 LIMA P ;P ;P ;14;14;14;14;4;2;2 Nuclear pore complex protein Nup214 NUP Q9Y394;H0YJ66;Q9Y394-2;H18;17;16;9;8;1 Dehydrogenase/reductase SDR family member 7 DHRS Q15125;C9JJ78;C9J719 2;1;1 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase EBP O43395;E7EVD1;B4DSY9 5;4;2 U4/U6 small nuclear ribonucleoprotein Prp3 PRPF Q9NX40;D6RBN5;G8JLN7;Q9;7;7;7;7;7;7;7;5;5;5;5OCIA domain-containing protein 1 OCIAD Q9P003;A6NLH6 1;1 Protein cornichon homolog 4 CNIH P U6 snrna-associated Sm-like protein LSm6 LSM Q9HCM4;Q9HCM4-3;Q9HC 2;2;2;2;1;1 Band 4.1-like protein 5;Band 4.1-like protein 4B EPB41L5;EPB41L4B P NEDD8-conjugating enzyme Ubc12 UBE2M Q9H9P8;C9JVN9;Q9H9P8-23;3;3;1;1 L-2-hydroxyglutarate dehydrogenase, mitochondrial L2HGDH O Torsin-1A TOR1A O75976;F5GZH6;J3QQJ4;J3Q42;34;7;1 Carboxypeptidase D CPD O95292;O ;E5RK64 10;3;3 Vesicle-associated membrane protein-associated proteinvapb P78357;K7EMM9 15;7 Contactin-associated protein 1 CNTNAP Q9NQC3-2;Q9NQC3;Q9NQ 10;9;9;9;6;6;5;5;5 Reticulon-4 RTN Q9NZE8;D3YTC1;Q9NZE8-2 2;1;1 39S ribosomal protein L35, mitochondrial MRPL Q9NQ50;F8WBK5 4;4 39S ribosomal protein L40, mitochondrial MRPL Q969E2;Q969E2-2;Q969E2-32;2;1;1;1 Secretory carrier-associated membrane protein 4 SCAMP P60953;E7ETU3;P ;Q10;8;8;7;1;1;1;1;1 Cell division control protein 42 homolog CDC Q99470;K7ELI9 4;1 Stromal cell-derived factor 2 SDF Q9NX20;E9PI14 8;4 39S ribosomal protein L16, mitochondrial MRPL REV H3BQK9;REV H3BP 3;3;3; J3KN66;Q5JTV8;H0Y4R4;E9 15;14;8;7;6;5;1 Torsin-1A-interacting protein 1 TOR1AIP O95071;J3KMW7;E7EMW7; 36;36;35;6;3;3;2 E3 ubiquitin-protein ligase UBR5 UBR Q Putative ATP-dependent Clp protease proteolytic subuniclpp P27105;B4E2V5;P ;5;1 Erythrocyte band 7 integral membrane protein STOM E7EQD5;Q15833;Q ;4;4 Syntaxin-binding protein 2 STXBP B5ME97;Q9P0V9-2;E7EX04;6;6;6;6;6;5;4;4;1;1;1;1Septin P20339;B4DJA5 9;8 Ras-related protein Rab-5A RAB5A P15907;H7C472;B2R513;C9J6;3;1;1;1;1 Beta-galactoside alpha-2,6-sialyltransferase 1 ST6GAL P ATP synthase subunit a MT-ATP Q9UJS0-2;Q9UJS0;F5GX33;O12;12;10;3;3 Calcium-binding mitochondrial carrier protein Aralar2 SLC25A Q9NYV4;Q9NYV4-2;Q9NYV 5;5;4;4 Cyclin-dependent kinase 12 CDK P78345;Q5VUC3;Q5VUC9 1;1;1 Ribonuclease P protein subunit p38 RPP38;RP11-455B Q8NFF5;Q8NFF5-2;Q8NFF55;5;5;3;2;1;1 FAD synthase;molybdenum cofactor biosynthesis proteinflad P83111;P ;H0YNN5 12;7;2 Serine beta-lactamase-like protein LACTB, mitochondriallactb Q ;E9PKU7;E9PNH1;54;44;7;5;2;1 GANAB P98160;Q5SZJ2;Q5SZJ1 53;4;1 Basement membrane-specific heparan sulfate proteogly HSPG P43304;P ;F8W6E4;F24;23;19;10 Glycerol-3-phosphate dehydrogenase, mitochondrial GPD Q969V3;Q969V3-2;K7EMW 8;8;7;4;3;2;1 Nicalin NCLN P09110;C9JDE9;H7C131;G5E14;12;11;9;6;4;3;2;2 3-ketoacyl-CoA thiolase, peroxisomal ACAA B7ZKM8;O95487;O ;5;5 Protein transport protein Sec24B SEC24B J3KPM8;K7EN75;P14854;K76;6;6;1 Cytochrome c oxidase subunit 6B1 COX6B O Barrier-to-autointegration factor BANF P Nuclear pore glycoprotein p62 NUP Q5BJF2;J3KTD1;Q86XC5;J3K4;3;2;1 Transmembrane protein 97 TMEM
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 04.02.2014 Αρ. πρωτ. 1054 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
ΜΟΡΙΑΚΕΣ ΜΕΘΟΔΟΙ ΚΡΙΤΗΡΙΑ ΕΠΙΛΟΓΗΣ ΑΞΙΟΛΟΓΗΣΗ
 - - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
- - ό ό : Ω ί ό ί ώ.. ά ή ί ός ή ς ύ ί ί ή έ ώ ό. ή ς ά ς ής ό ός ά ς ή έ ός ή ς ί ής ί ς. ό έ ώ - ώ ή ής ή ς- ί ά ά ς ί ς. ά ί ί έ έ ά ύ ή, ό ί ά, ό ό ά έ ά ής ί ύ George Wald ή ί έ ς ί ύ ό ς ί ς ά έ
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Supplementary Information Titles
 Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Supplementary Information Titles Journal: Nature Neuroscience Article Title: Corresponding Author: Wnt-mediated activation of NeuroD1 and retroelements during Tomoko Kuwabara adult neurogenesis Supplementary
Chapter 1. Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes
 Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Chapter 1 Fingolimod attenuates ceramide induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes 1 1 1 1 CHAPTER SUMMARY FINGOLIMOD ATTENUATES CERAMIDE PRODUCTION
Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου Cyld στον καρκίνο του μαστού
 Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
Σχολή Θετικών Επιστημών Τμήμα Βιολογίας Πρόγραμμα Μεταπτυχιακών Σπουδών Κατεύθυνση: Εφαρμοσμένη γενετική και βιοτεχνολογία ΜΕΤΑΠΤΥΧΙΑΚΗ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της έκφρασης του ογκοκατασταλτικού γονιδίου
ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ»
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΑΤΡΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ «ΚΛΙΝΙΚΕΣ ΚΑΙ ΚΛΙΝΙΚΟΕΡΓΑΣΤΗΡΙΑΚΕΣ ΙΑΤΡΙΚΕΣ ΕΙΔΙΚΟΤΗΤΕΣ» ΑΡΝΗΤΙΚΗ ΡΥΘΜΙΣΗ ΤΗΣ ΜΕΤΑΒΙΒΑΣΗΣ ΤΟΥ ΣΗΜΑΤΟΣ ΤΗΣ
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01
 SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
SABiosciences PCR Array Catalog #: PAHS-021 SA+ SCF 4h stimulaton AVG(Ct) Position Unigene Refseq Symbol Description shcontr shcontr-4h shgskβ A01 Hs.1274 NM_006129 BMP1 Bone morphogenetic protein 1 25.25
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Πρόσκληση υποβολής προσφοράς
 Αθήνα, 5 Μαρτίου 2014 Πρόσκληση υποβολής προσφοράς από φυσικά ή νομικά πρόσωπα για την προμήθεια υλικών στην Πράξη «ΘΑΛΗΣ Εθνικό Ίδρυμα Ερευνών Βιοσύνθεση και Γενετική Επιλογή Κυκλικών Πεπτιδίων με Εν
Αθήνα, 5 Μαρτίου 2014 Πρόσκληση υποβολής προσφοράς από φυσικά ή νομικά πρόσωπα για την προμήθεια υλικών στην Πράξη «ΘΑΛΗΣ Εθνικό Ίδρυμα Ερευνών Βιοσύνθεση και Γενετική Επιλογή Κυκλικών Πεπτιδίων με Εν
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary information:
 Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Supplementary information: Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging Shuai Guo 1, Dan Zhou
Figures Supplementary Figure 1. Tandem MS of yeast eif2b.
 Figures Supplementary Figure 1. Tandem MS of yeast eif2b. Tandem MS of the 56+ charge state (shaded blue) of the eif2b dimer showing that two α subunits ( at 2000 m/z) dissociate from the dimer when collision
Figures Supplementary Figure 1. Tandem MS of yeast eif2b. Tandem MS of the 56+ charge state (shaded blue) of the eif2b dimer showing that two α subunits ( at 2000 m/z) dissociate from the dimer when collision
Table S1: Inpatient Diet Composition
 Table S1: Inpatient Diet Composition Diet Components (% of Energy) Inpatient Period Baseline Intervention Protein 15.1 ± 0.0 15.0 ± 0.1 Total fat 30.0 ± 0.0 30.0 ± 0.1 Saturated fat 8.6 ± 0.8 8.4 ± 0.9
Table S1: Inpatient Diet Composition Diet Components (% of Energy) Inpatient Period Baseline Intervention Protein 15.1 ± 0.0 15.0 ± 0.1 Total fat 30.0 ± 0.0 30.0 ± 0.1 Saturated fat 8.6 ± 0.8 8.4 ± 0.9
High mobility group 1 HMG1
 Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Vol. 29, pp.705 ~ 711, 2001 High mobility group 1 HMG1 13 12 20 anti-neutrophil cytoplasmic antibodies, ANCA ANCA 1982 Davies 1980 1 high mobility group HMG1 HMG2 30 kd high mobility group HMGHMG HMG1
Structural and Biochemical Studies of Cotranslational Protein Transport across the Endoplasmic Reticulum
 Structural and Biochemical Studies of Cotranslational Protein Transport across the Endoplasmic Reticulum Inaugural-Dissertation zur Erlangung des akademischen Grades Doktor der Naturwissenschaften - Dr.
Structural and Biochemical Studies of Cotranslational Protein Transport across the Endoplasmic Reticulum Inaugural-Dissertation zur Erlangung des akademischen Grades Doktor der Naturwissenschaften - Dr.
Supporting Information
 Supporting Information Mitochondria-Targeting Polydopamine Nanocomposites as Chemophotothermal Therapeutics for Cancer Zhuo Wang *,, Yuzhi Chen, Hui Zhang, Yawen Li, Yufan Ma, Jia Huang, Xiaolei Liu, Fang
Supporting Information Mitochondria-Targeting Polydopamine Nanocomposites as Chemophotothermal Therapeutics for Cancer Zhuo Wang *,, Yuzhi Chen, Hui Zhang, Yawen Li, Yufan Ma, Jia Huang, Xiaolei Liu, Fang
Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ ΩΝ ΚΑΤΑ ΤΗΝ ΕΜΒΡΥΪΚΗ ΑΝΑΠΤΥΞΗ
 ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ Πρόγραµµα Μεταπτυχιακών Σπουδών Εφαρµογές στις Βασικές Ιατρικές Επιστήµες Εργαστήριο Ανατοµικής-Ιστολογίας-Εµβρυολογίας ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Ο ΡΟΛΟΣ ΤΩΝ ΚΑΝΝΑΒΙΝΟΕΙ
SUPPLEMENTARY INFORMATION
 Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Sensitivity of [Ru(phen) 2 dppz] 2+ Light Switch Emission to Ionic Strength, Temperature, and DNA Sequence and Conformation Andrew W. McKinley, Per Lincoln and Eimer M. Tuite* SUPPLEMENTARY INFORMATION
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor
 Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor SYDNEY P. THOMAS, 1 TRISH T. HOANG, 2 VALERIE T. RESSLER, 4 and RONALD T. RAINES 2,3,4 1 Graduate Program in Cell & Molecular
NOVEL INTEGRAL MEMBRANE PROTEINS OF THE INNER NUCLEAR MEMBRANE
 NOVEL INTEGRAL MEMBRANE PROTEINS OF THE INNER NUCLEAR MEMBRANE CHARACTERIZATION OF LUMA NATIVE LAP 2β COMPLEXES Inauguraldissertation zur Erlangung der Doktorwürde des Fachbereichs Biologie Chemie Pharmazie
NOVEL INTEGRAL MEMBRANE PROTEINS OF THE INNER NUCLEAR MEMBRANE CHARACTERIZATION OF LUMA NATIVE LAP 2β COMPLEXES Inauguraldissertation zur Erlangung der Doktorwürde des Fachbereichs Biologie Chemie Pharmazie
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ. ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥ ΩΝ «Ιατρική Ερευνητική Μεθοδολογία» ΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Νόσος Pompe: Νεότερα κλινικά, γενετικά, διαγνωστικά και θεραπευτικά
Aluminum Electrolytic Capacitors
 Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Aluminum Electrolytic Capacitors Snap-In, Mini., 105 C, High Ripple APS TS-NH ECE-S (G) Series: TS-NH Features Long life: 105 C 2,000 hours; high ripple current handling ability Wide CV value range (47
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Διαμόρφωση υποστρωμάτων στη μικροκαι νανο-κλίμακα για την δημιουργία πρωτεϊνικών μικροσυστοιχιών
 Διαμόρφωση υποστρωμάτων στη μικροκαι νανο-κλίμακα για την δημιουργία πρωτεϊνικών μικροσυστοιχιών Επιστημονικοί Υπεύθυνοι έργου: A. Τσερέπη Ινστιτούτο Μικροηλεκτρονικής Π. Σ. Πέτρου Ινστιτούτο Ραδιοϊσοτόπων
Διαμόρφωση υποστρωμάτων στη μικροκαι νανο-κλίμακα για την δημιουργία πρωτεϊνικών μικροσυστοιχιών Επιστημονικοί Υπεύθυνοι έργου: A. Τσερέπη Ινστιτούτο Μικροηλεκτρονικής Π. Σ. Πέτρου Ινστιτούτο Ραδιοϊσοτόπων
Synthesis and Biological Evaluation of Novel Acyclic and Cyclic Glyoxamide derivatives as Bacterial Quorum Sensing and Biofilm Inhibitors
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2017 Synthesis and Biological Evaluation of Novel Acyclic and Cyclic Glyoxamide
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2017 Synthesis and Biological Evaluation of Novel Acyclic and Cyclic Glyoxamide
Aluminum Electrolytic Capacitors (Large Can Type)
 Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Aluminum Electrolytic Capacitors (Large Can Type) Snap-In, 85 C TS-U ECE-S (U) Series: TS-U Features General purpose Wide CV value range (33 ~ 47,000 µf/16 4V) Various case sizes Top vent construction
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
CPT. Tsuchiya. beta. quantitative RT PCR QIAGEN IGFBP. Fect Transfection Reagent sirna. RT PCR RNA Affymetrix GeneChip Expression Array
 7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
7 PTEN p Tsuchiya DNA Hepatocyte nuclear factor beta HNF beta HNFbeta IGFBP GLUT G Pase MODY maturity onset diabetes of the young Tsuchiya HNFbeta HNFbeta HNFbeta sirna CPT HNFbeta HNFbeta TOVG KOC ESMCAS
Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance
 POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
POJ 7(1):19-27 (2014) ISSN:1836-3644 Overlapping sets of transcripts from host and non-host interactions of tomato are expressed early during non-host resistance Battepati Uma and Appa Rao Podile* Supplementary
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών
 Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Ζώων για Παραγωγή Προϊόντων Ποιότητας» 1 Αξιοποίηση Φυσικών Αντιοξειδωτικών στην Εκτροφή των Αγροτικών Ζώων για Παραγωγή Προϊόντων Ποιότητας Γεωπονικό Πανεπιστήμιο Αθηνών Εργαστήριο Ζωοτεχνίας MIS 380231
Supporting Information. Experimental section
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information Experimental section General. Proton nuclear magnetic resonance ( 1
Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks. Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks
 Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells
 Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee
Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells Yadavalli S. 1, Jayaram S. 1,2, Manda SS. 1,3, Madugundu AK. 1,3, Nayakanti DS. 1, Tuan TZ. 6, Bhat R. 4, Rangarajan A. 4, Chatterjee
Si + Al Mg Fe + Mn +Ni Ca rim Ca p.f.u
 .6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
.6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver.
 Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
Supplementary figures for MED13 dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. Content: Figure S1. Cardiac overexpression of MED13 increases metabolic
5- CACGAAACTACCTTCAACTCC-3 beta actin-r 5- CATACTCCTGCTTGCTGATC-3 GAPDH-F GAPDH-R
 Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΗΛΕΚΤΡΟΜΑΓΝΗΤΙΚΩΝ ΕΦΑΡΜΟΓΩΝ ΗΛΕΚΤΡΟΟΠΤΙΚΗΣ & ΗΛΕΚΤΡΟΝΙΚΩΝ ΥΛΙΚΩΝ Μελέτη Επίδρασης Υπεριώδους Ακτινοβολίας σε Λεπτά
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΒΙΟΧΗΜΕΙΑΣ & ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test Επιμέλεια
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΒΙΟΧΗΜΕΙΑΣ & ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ Μελέτη της αντιμεταλλαξιγόνου δράσης φλαβονοειδών του φυτού Lotus Edulis με τη μέθοδο Ames test Επιμέλεια
Table S2. Sequences immunoprecipitating with anti-hif-1α
 Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
Genome-wide analysis of HIF DNA-bdg-supplemental material Table S2. Sequences immunoprecipitatg with anti-α The table gives the chromosomal co-ordates of each anti-α immunoprecipitated locus meetg criteria
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
Abstract... I. Zusammenfassung... II. 1 Aim of the work Introduction Short overview of Chinese hamster ovary cell lines...
 III Contents I II III Abstract... I Zusammenfassung... II Contents... III 1 Aim of the work... 1 2 Introduction... 2 2.1 Short overview of Chinese hamster ovary cell lines... 2 2.2 Unspecific mutations:
III Contents I II III Abstract... I Zusammenfassung... II Contents... III 1 Aim of the work... 1 2 Introduction... 2 2.1 Short overview of Chinese hamster ovary cell lines... 2 2.2 Unspecific mutations:
Supplementary Figure 1
 NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
rp, ribosomal protein 60S rp mrna rpl30 rps14 RT-PCR mrna nonsense-mediated mrna decay smg-2 smg-2 mrna rp rpl-1 rpl-1
 rp, ribosomal protein 60S 47 rpl 40S 32 rps rp mrna rpl30 rps14 rpl12 rpl3 rps13 rp mrna nonsense-mediated mrna decay (NMD) C. elegans smg-2 smg-2 mrna 50 rp 7 rp 4 rpl 4 rp NMD mrna 6 rp mrna rp mrna
rp, ribosomal protein 60S 47 rpl 40S 32 rps rp mrna rpl30 rps14 rpl12 rpl3 rps13 rp mrna nonsense-mediated mrna decay (NMD) C. elegans smg-2 smg-2 mrna 50 rp 7 rp 4 rpl 4 rp NMD mrna 6 rp mrna rp mrna
Science of Sericulture
 Science of Sericulture 2012 38 1 0065-0069 ISSN 0257-4799 CN 32-1115 /S E-mail CYKE@ chinajournal. net. cn 215123 25 0 ~ 24 h GSH GSSG GSH + 2GSSG GSH /GSSG S- GST TPX 23% GR 57% GSH TPX GST GSSG 61% GSH
Science of Sericulture 2012 38 1 0065-0069 ISSN 0257-4799 CN 32-1115 /S E-mail CYKE@ chinajournal. net. cn 215123 25 0 ~ 24 h GSH GSSG GSH + 2GSSG GSH /GSSG S- GST TPX 23% GR 57% GSH TPX GST GSSG 61% GSH
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to. facilitate intracellular survival
 Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Salmonella produce microrna-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival Hongwei Gu 1,2*, Chihao Zhao 1,2*, Tianfu Zhang 1,2*, Hongwei Liang 1,2, Xiao-Ming Wang 1,
Chalkou I. C. [PROJECT] Ανάθεση εργασιών.
![Chalkou I. C. [PROJECT] Ανάθεση εργασιών. Chalkou I. C. [PROJECT] Ανάθεση εργασιών.](/thumbs/29/13219375.jpg) Πληροφορική της Υγείας 2014 Chalkou I. C. [PROJECT] Ανάθεση εργασιών. Περιεχόμενα 1. Ομάδα ΣΤ... 3 1.1 ΜΑΡΚΟΠΟΥΛΟΥ- ΣΠΥΡΟΠΟΥΛΟΥ -ΚΩΝΣΤΑΝΤΟΠΟΥΛΟΥ... 3 1.2 ΜΑΡΚΟΣ- ΚΟΥΤΣΟΠΟΥΛΟΣ ΑΥΓΕΡΗ - ΜΠΟΥΖΑΛΑ... 3 1.3
Πληροφορική της Υγείας 2014 Chalkou I. C. [PROJECT] Ανάθεση εργασιών. Περιεχόμενα 1. Ομάδα ΣΤ... 3 1.1 ΜΑΡΚΟΠΟΥΛΟΥ- ΣΠΥΡΟΠΟΥΛΟΥ -ΚΩΝΣΤΑΝΤΟΠΟΥΛΟΥ... 3 1.2 ΜΑΡΚΟΣ- ΚΟΥΤΣΟΠΟΥΛΟΣ ΑΥΓΕΡΗ - ΜΠΟΥΖΑΛΑ... 3 1.3
Supplementary Table 1. Construct List with key Biophysical Properties of the expression
 SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
SPINE Benchmark Target ID Well Code Tag (N or C) Fusion MW (kda) Fusion pi Cleavage site Prot MW (Da) Prot pi 1 A1 OPPF 2585 N-His6 21.75 6.41 Protease 3C 19.77 5.43 2 B1 OPPF 2586 N-His6 15.6 6.27 Protease
Guo_Fig. S1. Atg7 +/+ Atg7 -/- FBP_M+6 DHAP_M+3. Glucose Uptake Rate G6P_M+6. nmol/ul cell/hr
 Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Guo_Fig. S1 A C D nmol/ul cell/hr nmol/ul cell/hr Glucose Uptake Rate.3.2.1 *. Lactate Secretion Rate.4.3.2.1. Atg7-/- B Glucose G3P Lac Glucose G6P FBP 3-PG Pyr DHAP cycle Carbon from labeled glucose
Sar1B-mediated Regulation of Triglyceride and Cholesterol
 FIGURE S1. Comparing Sar1A- and Sar1B-flag Over-expression on RNA Profiles of McArdle RH7777 cells. RNA was prepared from two Sar1B:H79G-flag cell lines and two control cell lines plus two Sar1B-flag and
FIGURE S1. Comparing Sar1A- and Sar1B-flag Over-expression on RNA Profiles of McArdle RH7777 cells. RNA was prepared from two Sar1B:H79G-flag cell lines and two control cell lines plus two Sar1B-flag and
Acknowledgements... 3 Contents... I Figures... VII Tables... IX Abbreviations... X
 I Acknowledgements... 3... I Figures... VII Tables... IX Abbreviations... X Amino acids... XII Nucleobases... XII 1 Introduction... 1 1.1 Polyketides and non-ribosomal peptides... 1 1.2 Polyketide synthases...
I Acknowledgements... 3... I Figures... VII Tables... IX Abbreviations... X Amino acids... XII Nucleobases... XII 1 Introduction... 1 1.1 Polyketides and non-ribosomal peptides... 1 1.2 Polyketide synthases...
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver
 Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Nagoya Med. J., 137 Hepatic Stellate Cells: Multifunctional mesenchymal cells of the Liver KAZUO IKEDA Department of Anatomy and Cell Biology, Graduate School of Medical Sciences, Nagoya City University
Table S1A. Generic EMT signature for tumour
 Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
(1) Describe the process by which mercury atoms become excited in a fluorescent tube (3)
 Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
Q1. (a) A fluorescent tube is filled with mercury vapour at low pressure. In order to emit electromagnetic radiation the mercury atoms must first be excited. (i) What is meant by an excited atom? (1) (ii)
w o = R 1 p. (1) R = p =. = 1
 Πανεπιστήµιο Κρήτης - Τµήµα Επιστήµης Υπολογιστών ΗΥ-570: Στατιστική Επεξεργασία Σήµατος 205 ιδάσκων : Α. Μουχτάρης Τριτη Σειρά Ασκήσεων Λύσεις Ασκηση 3. 5.2 (a) From the Wiener-Hopf equation we have:
Πανεπιστήµιο Κρήτης - Τµήµα Επιστήµης Υπολογιστών ΗΥ-570: Στατιστική Επεξεργασία Σήµατος 205 ιδάσκων : Α. Μουχτάρης Τριτη Σειρά Ασκήσεων Λύσεις Ασκηση 3. 5.2 (a) From the Wiener-Hopf equation we have:
Μαρία Κατσιφοδήμου. Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα έμβρυα στην επιτυχία της εξωσωματικής γονιμοποίησης. Μεταπτυχιακή Διπλωματική Εργασία
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΘΕΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΒΙΟΛΟΓΙΑΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΚΑΤΕΥΘΥΝΣΗ: ΕΦΑΡΜΟΣΜΕΝΗ ΓΕΝΕΤΙΚΗ ΚΑΙ ΒΙΟΤΕΧΝΟΛΟΓΙΑ Ο ρόλος της έκκρισης HLA-G από τα ανθρώπινα
Supporting Information
 Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were
Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were
Supporting Information. Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka
 Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
Supporting Information Multigenerational Disruption of Thyroid Endocrine System in Marine Medaka after A Life-Cycle Exposure to Perfluorobutane Sulfonate (PFBS) Lianguo Chen,, Chenyan Hu #, Mirabelle M.
ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ
 1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 12/06/2013 Αρ. πρωτ. 4972 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
1 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΕΙΔΙΚΟΣ ΛΟΓΑΡΙΑΣΜΟΣ ΑΝΑΡΤΗΤΕΑ ΣΤΟ ΔΙΑΔΙΚΤΥΟ ΠΡΟΣΚΛΗΣΗ ΕΚΔΗΛΩΣΗΣ ΕΝΔΙΑΦΕΡΟΝΤΟΣ Ηράκλειο, 12/06/2013 Αρ. πρωτ. 4972 Ο Ειδικός Λογαριασμός του Πανεπιστημίου Κρήτης
TGFp FSH INH INH
 (210095) E-mail ( genlinwang@hotmail.com) - -(Strept Avidin-Biotin-Peroxidase Complex SABC), 1 32,, 1 inhibin INH α 32 000 TGFp FSH FSH [1] INHα Sertoli Noguchi 1997 [2] Meunier et al.1988 [3] INH INH
(210095) E-mail ( genlinwang@hotmail.com) - -(Strept Avidin-Biotin-Peroxidase Complex SABC), 1 32,, 1 inhibin INH α 32 000 TGFp FSH FSH [1] INHα Sertoli Noguchi 1997 [2] Meunier et al.1988 [3] INH INH
Journal of Ningxia Medical University. Annexin P19 mrna P < AnnexinA2 P19 mrna. AnnexinA2 P19 mrna. PCR 1.
 33 6 2011 6 Journal of Ningxia Medical University 511 1674-6309 2011 06-0511 - 04 AnnexinA2 P19 1 2 3 3 3 1. 415700 2. 750004 3. 750004 AnnexinA2 P19 62 IHC RT - PCR Western blot AnnexinA2P19 mrna AnnexinA2
33 6 2011 6 Journal of Ningxia Medical University 511 1674-6309 2011 06-0511 - 04 AnnexinA2 P19 1 2 3 3 3 1. 415700 2. 750004 3. 750004 AnnexinA2 P19 62 IHC RT - PCR Western blot AnnexinA2P19 mrna AnnexinA2
IL - 13 /IL - 18 ELISA PCR RT - PCR. IL - 13 IL - 18 mrna. 13 IL - 18 mrna IL - 13 /IL Th1 /Th2
 344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
344 IL - 13 /IL - 18 1 2 1 2 1 2 1 2 1 2 3 1 2 13 18 IL - 13 /IL - 18 10% / OVA /AL OH 3 5% 16 ~ 43 d 44 d ELISA BALF IL - 13 IL - 18 PCR RT - PCR IL - 13 IL - 18 mrna IL - 13 mrna 0. 01 IL - 18 mrna 0.
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ. Πτυχιακή εργασία
 ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
ΤΕΧΝΟΛΟΓΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΥΠΡΟΥ ΣΧΟΛΗ ΓΕΩΠΟΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΚΑΙ ΕΠΙΣΤΗΜΗΣ ΤΡΟΦΙΜΩΝ Πτυχιακή εργασία ΜΕΛΕΤΗ ΠΟΛΥΦΑΙΝΟΛΩΝ ΚΑΙ ΑΝΤΙΟΞΕΙΔΩΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΣΟΚΟΛΑΤΑΣ Αναστασία Σιάντωνα Λεμεσός
Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ ΡΑΣΗ ΤΟΥ ΓΑΣΤΡΙΚΟΥ ΒΛΕΝΝΟΓΟΝΟΥ ΣΤΗ ΛΟΙΜΩΞΗ ΜΕ ΕΛΙΚΟΒΑΚΤΗΡΙ ΙΟ ΤΟΥ ΠΥΛΩΡΟΥ ΠΡΙΝ ΚΑΙ ΜΕΤΑ ΤΗ ΘΕΡΑΠΕΙΑ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΤΟΜΕΑΣ ΥΓΕΙΑΣ ΕΡΓΑΣΤΗΡΙΑΚΟΣ ΙΕΥΘΥΝΤΗΣ: Ο ΚΑΘΗΓΗΤΗΣ ΓΕΩΡΓΙΟΣ ΚΑΡΚΑΒΕΛΑΣ ΠΑΝΕΠ. ΕΤΟΣ 2008-2009 Αριθµ. 2084 Η ΦΛΕΓΜΟΝΩ ΗΣ ΑΝΤΙ
Hvordan påvirker CO 2 post-smolt i brakkvann RAS?
 Hvordan påvirker CO 2 post-smolt i brakkvann RAS? Tom O. Nilsen,. Mota, V.C., Kolarevic, J., Ytterborg, E., Ebbesson, Lars O.E., Handeland, S.O., Stiller, K., Terjesen, B.F SUNNDALSØRA, 12. NOVEMBER 2018
Hvordan påvirker CO 2 post-smolt i brakkvann RAS? Tom O. Nilsen,. Mota, V.C., Kolarevic, J., Ytterborg, E., Ebbesson, Lars O.E., Handeland, S.O., Stiller, K., Terjesen, B.F SUNNDALSØRA, 12. NOVEMBER 2018
BD Cytometric Bead Array Flex Set System
 BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
BD Cytometric Bead Array Flex Set System BD Cytometric Bead Array System BD Biosciences Pharmingen ELISA Cytometric Bead Array CBA System Capture Caputure Serum Supernatant Lysate PE -Detection 1 2 PE
http / / cjbmb. bjmu. edu. cn Chinese Journal of Biochemistry and Molecular Biology HBx HBxΔ127
 ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 5 27 5 426 ~ 430 X HBxΔ127 ERK 1 * * 300071 hepatitis B virus HBV X HBx HBx HBxΔ127 1
ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 5 27 5 426 ~ 430 X HBxΔ127 ERK 1 * * 300071 hepatitis B virus HBV X HBx HBx HBxΔ127 1
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
ΔΙΑΣΤΑΣΕΙΣ ΕΣΩΤΕΡΙΚΗΣ ΓΩΝΙΑΣ INTERNAL CORNER SIZES
 ΔΙΑΣΤΑΣΕΙΣ ΕΣΩΤΕΡΙΚΗΣ ΓΩΝΙΑΣ 90 90 INTERNAL CORNER SIZES ΟΠΤΙΚΗ PERSPECTIVE ΠΑΝΩ ΟΨΗ TOP VIEW ΔΙΑΣΤΑΣΕΙΣ ΡΑΦΙΩΝ SHELF DIMENSIONS T1 ΜΕΓΙΣΤΟ ΕΠΙΤΡΕΠΟΜΕΝΟ ΦΟΡΤΙΟ (1) MAXIMUM LOADING CAPACITIES (1) ΤΥΠΙΚΑ
ΔΙΑΣΤΑΣΕΙΣ ΕΣΩΤΕΡΙΚΗΣ ΓΩΝΙΑΣ 90 90 INTERNAL CORNER SIZES ΟΠΤΙΚΗ PERSPECTIVE ΠΑΝΩ ΟΨΗ TOP VIEW ΔΙΑΣΤΑΣΕΙΣ ΡΑΦΙΩΝ SHELF DIMENSIONS T1 ΜΕΓΙΣΤΟ ΕΠΙΤΡΕΠΟΜΕΝΟ ΦΟΡΤΙΟ (1) MAXIMUM LOADING CAPACITIES (1) ΤΥΠΙΚΑ
Highly enantioselective cascade synthesis of spiropyrazolones. Supporting Information. NMR spectra and HPLC traces
 Highly enantioselective cascade synthesis of spiropyrazolones Alex Zea a, Andrea-Nekane R. Alba a, Andrea Mazzanti b, Albert Moyano a and Ramon Rios a,c * Supporting Information NMR spectra and HPLC traces
Highly enantioselective cascade synthesis of spiropyrazolones Alex Zea a, Andrea-Nekane R. Alba a, Andrea Mazzanti b, Albert Moyano a and Ramon Rios a,c * Supporting Information NMR spectra and HPLC traces
Η Α ΕΝΟΫΠΟΦΥΣΗ: OI ΓΟΝΑ ΟΤΡΟΠΙΝΕΣ (FSH, LH) ΚΑΙ Η ΠΡΟΛΑΚΤΙΝΗ (PRL)
 Η Α ΕΝΟΫΠΟΦΥΣΗ: OI ΓΟΝΑ ΟΤΡΟΠΙΝΕΣ (FSH, LH) ΚΑΙ Η ΠΡΟΛΑΚΤΙΝΗ (PRL) Κωνσταντίνος Καλλαράς Ιατρός Παθολόγος Καθηγητής Φυσιολογίας Εργαστήριο Πειραματικής Φυσιολογίας Ιατρικής Σχολής Α.Π.Θ. FSH:ΜΒ 33000,
Η Α ΕΝΟΫΠΟΦΥΣΗ: OI ΓΟΝΑ ΟΤΡΟΠΙΝΕΣ (FSH, LH) ΚΑΙ Η ΠΡΟΛΑΚΤΙΝΗ (PRL) Κωνσταντίνος Καλλαράς Ιατρός Παθολόγος Καθηγητής Φυσιολογίας Εργαστήριο Πειραματικής Φυσιολογίας Ιατρικής Σχολής Α.Π.Θ. FSH:ΜΒ 33000,
Supporting Information
 Supporting Information Aluminum Complexes of N 2 O 2 3 Formazanate Ligands Supported by Phosphine Oxide Donors Ryan R. Maar, Amir Rabiee Kenaree, Ruizhong Zhang, Yichen Tao, Benjamin D. Katzman, Viktor
Supporting Information Aluminum Complexes of N 2 O 2 3 Formazanate Ligands Supported by Phosphine Oxide Donors Ryan R. Maar, Amir Rabiee Kenaree, Ruizhong Zhang, Yichen Tao, Benjamin D. Katzman, Viktor
J. Dairy Sci. 93: doi: /jds American Dairy Science Association, 2010.
 Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
Supplementary Table 1. Primers and PCR conditions used for the amplification of the goat SCD1 cdna (PCR1 to PCR6) and three SCD1 polymorphic regions (PCR7 to PCR9) PCR Primers Sequence Position 1 Thermal
the total number of electrons passing through the lamp.
 1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
1. A 12 V 36 W lamp is lit to normal brightness using a 12 V car battery of negligible internal resistance. The lamp is switched on for one hour (3600 s). For the time of 1 hour, calculate (i) the energy
Strain gauge and rosettes
 Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Supporting Information
 Supporting Information Wiley-VCH 27 69451 Weinheim, Germany Supplementary Figure 1. Synthetic results as detected by XRD (Cu-Kα). Simulation pattern of MCM-68 Relative Intensity / a.u. YNU-2P Conventional
Supporting Information Wiley-VCH 27 69451 Weinheim, Germany Supplementary Figure 1. Synthetic results as detected by XRD (Cu-Kα). Simulation pattern of MCM-68 Relative Intensity / a.u. YNU-2P Conventional
ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ
 ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΚΑΛΛΙΟΠΗ ΚΩΤΣΑ ΕΠΙΚ. ΚΑΘΗΓΗΤΡΙΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ Α.Π.Θ ΤΜΗΜΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ-ΔΙΑΒΗΤΟΛΟΓΙΚΟ
ΚΥΤΤΑΡΙΚΟΣ ΘΑΝΑΤΟΣ ΚΑΙ ΑΝΑΓΕΝΝΗΣΗ Β-ΚΥΤΤΑΡΟΥ ΝΕΟΤΕΡΑ ΔΕΔΟΜΕΝΑ ΓΙΑ ΤΗ ΘΕΡΑΠΕΙΑ ΤΟΥ ΣΑΚΧΑΡΩΔΗ ΔΙΑΒΗΤΗ ΚΑΛΛΙΟΠΗ ΚΩΤΣΑ ΕΠΙΚ. ΚΑΘΗΓΗΤΡΙΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ ΙΑΤΡΙΚΗ ΣΧΟΛΗ Α.Π.Θ ΤΜΗΜΑ ΕΝΔΟΚΡΙΝΟΛΟΓΙΑΣ-ΔΙΑΒΗΤΟΛΟΓΙΚΟ
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3
 Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Lecture 2: Dirac notation and a review of linear algebra Read Sakurai chapter 1, Baym chatper 3 1 State vector space and the dual space Space of wavefunctions The space of wavefunctions is the set of all
Electrolyzed-Reduced Water as Artificial Hot Spring Water
 /-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
/-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
Η ΤΟΞΙΚΟΤΗΤΑ ΤΟΥ ΒΟΡΙΟΥ(B) ΣΤΗΝ ΚΑΛΛΙΕΡΓΕΙΑ ΤΗΣ ΤΟΜΑΤΑΣ
 ΑΛΕΞΑΝΔΡΕΙΟ Τ.Ε.Ι. ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΑΣ ΓΕΩΠΟΝΙΑΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΚΑΙ ΔΙΑΤΡΟΦΗΣ ΤΜΗΜΑ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΑΝΑΤΟΜΙΑΣ ΚΑΙ ΦΥΣΙΟΛΟΓΙΑΣ ΦΥΤΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η ΤΟΞΙΚΟΤΗΤΑ ΤΟΥ ΒΟΡΙΟΥ(B)
ΑΛΕΞΑΝΔΡΕΙΟ Τ.Ε.Ι. ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΤΕΧΝΟΛΟΓΙΑΣ ΓΕΩΠΟΝΙΑΣ ΚΑΙ ΤΕΧΝΟΛΟΓΙΑΣ ΤΡΟΦΙΜΩΝ ΚΑΙ ΔΙΑΤΡΟΦΗΣ ΤΜΗΜΑ ΦΥΤΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΑΝΑΤΟΜΙΑΣ ΚΑΙ ΦΥΣΙΟΛΟΓΙΑΣ ΦΥΤΩΝ ΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ Η ΤΟΞΙΚΟΤΗΤΑ ΤΟΥ ΒΟΡΙΟΥ(B)
Applications. 100GΩ or 1000MΩ μf whichever is less. Rated Voltage Rated Voltage Rated Voltage
 Features Rated Voltage: 100 VAC, 4000VDC Chip Size:,,,,, 2220, 2225 Electrical Dielectric Code EIA IEC COG 1BCG Applications Modems LAN / WAN Interface Industrial Controls Power Supply Back-Lighting Inverter
Features Rated Voltage: 100 VAC, 4000VDC Chip Size:,,,,, 2220, 2225 Electrical Dielectric Code EIA IEC COG 1BCG Applications Modems LAN / WAN Interface Industrial Controls Power Supply Back-Lighting Inverter
Multilayer Ceramic Chip Capacitors
 FEATURES X7R, X6S, X5R AND Y5V DIELECTRICS HIGH CAPACITANCE DENSITY ULTRA LOW ESR & ESL EXCELLENT MECHANICAL STRENGTH NICKEL BARRIER TERMINATIONS RoHS COMPLIANT SAC SOLDER COMPATIBLE* PART NUMBER SYSTEM
FEATURES X7R, X6S, X5R AND Y5V DIELECTRICS HIGH CAPACITANCE DENSITY ULTRA LOW ESR & ESL EXCELLENT MECHANICAL STRENGTH NICKEL BARRIER TERMINATIONS RoHS COMPLIANT SAC SOLDER COMPATIBLE* PART NUMBER SYSTEM
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
 1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Ενδοκυττάρια ιαµερίσµατα, ιαλογή και µεταφορά πρωτεινών
 Ενδοκυττάρια ιαµερίσµατα, ιαλογή και µεταφορά πρωτεινών Ένα ηπατικό κύτταρο σε διατοµή Η.Μ Compartment Main Function Cytosol contains many metabolic pathways (Chapters 3 and 4); protein synthesis (Chapter
Ενδοκυττάρια ιαµερίσµατα, ιαλογή και µεταφορά πρωτεινών Ένα ηπατικό κύτταρο σε διατοµή Η.Μ Compartment Main Function Cytosol contains many metabolic pathways (Chapters 3 and 4); protein synthesis (Chapter
I ΙΑΙΤΕΡΟΤΗΤΕΣ ΤΗΣ ΙΑΠΙΣΤΕΥΣΗΣ ΤΩΝ ΜΙΚΡΟΒΙΟΛΟΓΙΚΩΝ ΕΡΓΑΣΤΗΡΙΩΝ ΝΕΡΟΥ ΚΑΤΑ ISO 17025. ΠΗΓΕΣ ΑΝΟΜΟΙΟΓΕΝΕΙΑΣ ΚΑΙ ΛΑΘΩΝ
 I ΙΑΙΤΕΡΟΤΗΤΕΣ ΤΗΣ ΙΑΠΙΣΤΕΥΣΗΣ ΤΩΝ ΜΙΚΡΟΒΙΟΛΟΓΙΚΩΝ ΕΡΓΑΣΤΗΡΙΩΝ ΝΕΡΟΥ ΚΑΤΑ ISO 17025. Ρ. Αθηνά Μαυρίδου Καθ. Μικροβιολογίας ΤΕΙ Αθήνας ΠΗΓΕΣ ΑΝΟΜΟΙΟΓΕΝΕΙΑΣ ΚΑΙ ΛΑΘΩΝ Φυσική ανοµοιογένεια νερού στην προέλευση
I ΙΑΙΤΕΡΟΤΗΤΕΣ ΤΗΣ ΙΑΠΙΣΤΕΥΣΗΣ ΤΩΝ ΜΙΚΡΟΒΙΟΛΟΓΙΚΩΝ ΕΡΓΑΣΤΗΡΙΩΝ ΝΕΡΟΥ ΚΑΤΑ ISO 17025. Ρ. Αθηνά Μαυρίδου Καθ. Μικροβιολογίας ΤΕΙ Αθήνας ΠΗΓΕΣ ΑΝΟΜΟΙΟΓΕΝΕΙΑΣ ΚΑΙ ΛΑΘΩΝ Φυσική ανοµοιογένεια νερού στην προέλευση
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL 1. Methods MicroRNA TaqMan Low Density Array MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Life Technologies). Global
SUPPLEMENTAL MATERIAL 1. Methods MicroRNA TaqMan Low Density Array MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Life Technologies). Global
(MACF1) Association of Microtubule Actin Crosslinking Factor 1 with Cytoskeleton in Osteoblast
 13 5 Vol13 No5 2009 10 Life Science Research Oct 2009 2009 MACF1,,,,, * 710072 : (MACF1) (MG-63 MC3T3) /, MACF1,, MACF1 ; B MACF1,, MACF1 sirna-macf1 MG-63 MACF1, ; MACF1 : 1; ; ; ; : Q291 : A :1007-7847(2009)05-0382-06
13 5 Vol13 No5 2009 10 Life Science Research Oct 2009 2009 MACF1,,,,, * 710072 : (MACF1) (MG-63 MC3T3) /, MACF1,, MACF1 ; B MACF1,, MACF1 sirna-macf1 MG-63 MACF1, ; MACF1 : 1; ; ; ; : Q291 : A :1007-7847(2009)05-0382-06
Pentoxifylline ameliorated nonalcoholic fatty liver disease in the hyperglycemia and dyslipidemia mice by the up-regulation of fatty acid β-oxidation.
 Pentoxifylline ameliorated nonalcoholic fatty liver disease in the hyperglycemia and dyslipidemia mice by the up-regulation of fatty acid β-oxidation. Jia-Hung Ye 1, Jung Chao 2, Ming-Ling Chang 3, Wen-Huang
Pentoxifylline ameliorated nonalcoholic fatty liver disease in the hyperglycemia and dyslipidemia mice by the up-regulation of fatty acid β-oxidation. Jia-Hung Ye 1, Jung Chao 2, Ming-Ling Chang 3, Wen-Huang
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ
 ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΤΜΗΜΑ ΟΔΟΝΤΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΟΔΟΝΤΙΚΗΣ ΚΑΙ ΑΝΩΤΕΡΑΣ ΠΡΟΣΘΕΤΙΚΗΣ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ ΤΗΣ ΣΥΓΚΡΑΤΗΤΙΚΗΣ ΙΚΑΝΟΤΗΤΑΣ ΟΡΙΣΜΕΝΩΝ ΠΡΟΚΑΤΑΣΚΕΥΑΣΜΕΝΩΝ ΣΥΝΔΕΣΜΩΝ ΑΚΡΙΒΕΙΑΣ
