Predictive Models for Fast and Effective Profiling of Kinase Inhibitors
|
|
|
- Χλωρίς Αντωνιάδης
- 7 χρόνια πριν
- Προβολές:
Transcript
1 Predictive Models for Fast and Effective Profiling of Kinase Inhibitors Alina Bora, 1,2,# Sorin Avram, 2,#, * Ionel Ciucanu, 1 Marius Raica 3, Stefana Avram 4 1 Department of Chemistry, West University of Timisoara, Faculty of Chemistry -Biology-Geography, 16 Pestalozzi Str., , Timisoara, Romania 2 Department of Computational Chemistry, Institute of Chemistry Timisoara of Romanian Academy, 24 Mihai Viteazu Avenue, Timisoara, , Romania 3 Department of Microscopic Morphology/Histology, Angiogenesis Research Center, University of Medicine and Pharmacy Victor Babes Timisoara, 2 Eftimie Murgu, Timisoara, , Romania 4 Department Pharmacy II, Discipline of Pharmacognosy, University of Medicine and Pharmacy Victor Babes Timisoara, Faculty of # These authors contributed equally to the article. Pharmacy, 2 Eftimie Murgu, Timisoara, , Romania *Corresponding author. sorin_avram@acad-icht.tm.edu.ro or 5orin.4vram@gmail.com, Phone: Table S1. Description of kinase sets of actives... 2 Table S2. Standardization steps and parameters applied to process molecular structures using module Chemaxon Standardizer Table S3. Control parameters for random forest model generation using cforest, 6-9 in package party available in R statistical software system Table S4. Number of active and inactive compounds (according to the corresponding data source) in the training and test sets... 5 Table S5. Median, average and standard deviation of the number of compounds and the actives (computed per kinase set) ranked before the top 0.5% FP in the external VS test sets... 7 Table S6. Wilcoxon right shift p-values (α = 0.05): row elements contain significantly greater values compared to column elements; median values are reported in parentheses... 7 Table S7. Classification results of the kinase model prediction for kinase sets with actives and inactives defined as pk i 7 and pk i < 5, respectively, in Metz et al 11 profiling results... 8 Table S8. Classification results of the kinase model prediction for kinase sets with actives and inactives defined as scaled Activity % 0.1 and > 0.9, respectively, in Anastassiadis et al 12 profiling kinase sets Figure S1. Density distribution plots of the activity values (IC 50 in µm) and hot spot cutoff (red line) for the first three kinases (according to the ID number assigned in the paper) in cluster k1 (a), k2 (b), k3 (c) and k4 (d). The plot title contains the UniProt ID of the kinase and the ID number (in parenthesis) used in the paper Figure S2. Dendrograms resulted from Ward hierarchical clustering of 107 kinase sets in a first run (left), resulting in k4 (magenta) and a larger cluster of 103 kinase sets. These were clustered separately into 3 more groups (right) k2 (blue), k3 (red) and k1 (green) Figure S3. Accuracy (Acc), sensitivity (Se) and specificity (Sp) values of the internal (training sets) evaluation of the 107 kinase models Figure S4. Phylogenetic tree of the human kinases (generated using Kinome Render 13, 14 ) Target IDs and abbreviations of the kinases modeled in the current study are colored in red; two Atypical:PI3/PI4-kinase kinases, i.e., PIK3CA and PIK3CG could not be mapped and are not represented. Illustration reproduced courtesy of Cell Signaling Technology, Inc. ( 13 Figure S5. The distributions of (the kinase-set values) eroce (a, performed on the VS test sets) and AUC (b, performed on the classification test sets) according to the nine kinase groups Figure S6. Venn plot (created using the online software from Microarray Center CRP-Sante) 15 of the chemical scaffolds (BMFs) contained in the PubChemKinIna (only inactives), ChEMBL (only kinase inhibitors) and ColBioS-FlavRC 16 datasets
2 Table S1. Description of kinase sets of actives ID 1 Uniprot ID 2 GENE Name Kinase group Kinase Cluster Num of Actives Num of BMFs 3 Actives /BMF 4 avg TanD 5 avg IC 50 6 Hotspot/ IC 50 cutoff 7 1 P35968 KDR TK / P00533 EGFR TK / Q16539 MAPK14 CMGC / P42336 PIK3CA Atypical / P12931 SRC TK / P08581 MET TK /0.4 7 P49841 GSK3B CMGC / O14757 CHEK1 CAMK / O14965 AURKA Other / P04626 ERBB2 TK / P06239 LCK TK / P31749 AKT1 AGC / P48736 PIK3CG Atypical / P42345 MTOR Atypical / P24941 CDK2 CMGC / P08069 IGF1R TK / P45983 MAPK8 CMGC / P06493 CDK1 CMGC / O14920 IKBKB Other / O60674 JAK2 TK / Q96GD4 AURKB Other / P36888 FLT3 TK / P17948 FLT1 TK / P09619 PDGFRB TK / Q02763 TEK TK / P10721 KIT TK / P53779 MAPK10 CMGC / P52333 JAK3 TK /1 29 P11362 FGFR1 TK / O75116 ROCK2 AGC / P00519 ABL1 TK / P11309 PIM1 CAMK / Q08881 ITK TK / P45984 MAPK9 CMGC / Q02750 MAP2K1 STE / P15056 BRAF TKL / P43405 SYK TK /1 38 P04049 RAF1 TKL / Q13464 ROCK1 AGC / Q9P1W9 PIM2 CAMK / O00311 CDC7 Other / P16234 PDGFRA TK / P04629 NTRK1 TK / Q02156 PRKCE AGC /1.3 2
3 45 P49840 GSK3A CMGC / P35916 FLT4 TK / Q5S007 LRRK2 TKL / Q86V86 PIM3 CAMK / P29597 TYK2 TK / P78527 PRKDC Atypical / P49137 MAPKAPK2 CAMK /3 52 P36897 TGFBR1 TKL / P06213 INSR TK / P54760 EPHB4 TK / P17252 PRKCA AGC / P11802 CDK4 CMGC / O15111 CHUK Other / Q05397 PTK2 TK / Q13627 DYRK1A CMGC / P23458 JAK1 TK / P07949 RET TK / P51812 RPS6KA3 AGC / P30530 AXL TK /2 64 P43403 ZAP70 TK / P49759 CLK1 CMGC / P48730 CSNK1D CK / Q15759 MAPK11 CMGC / Q9NWZ3 IRAK4 TKL / Q00535 CDK5 CMGC / P28482 MAPK1 CMGC / Q9HAZ1 CLK4 CMGC / Q13557 CAMK2D CAMK / P51955 NEK2 Other / Q13535 ATR Atypical / P06241 FYN TK / P50613 CDK7 CMGC / Q9UM73 ALK TK / Q05655 PRKCD AGC / P53350 PLK1 Other / Q04759 PRKCQ AGC / P31751 AKT2 AGC / P05771 PRKCB AGC / P08631 HCK TK / O96017 CHEK2 CAMK / O15530 PDPK1 AGC / P07948 LYN TK / Q06418 TYRO3 TK / P24723 PRKCH AGC / P41279 MAP3K8 STE / Q06187 BTK TK / Q9Y243 AKT3 AGC / P05129 PRKCG AGC /2 93 Q12866 MERTK TK /0.23 3
4 94 P33981 TTK Other / Q9Y463 DYRK1B CMGC / Q9UHD2 TBK1 Other / Q13882 PTK6 TK / Q15139 PRKD1 CAMK / Q07912 TNK2 TK / Q9H4B4 PLK3 Other / Q9NYY3 PLK2 Other / Q14289 PTK2B TK / Q14164 IKBKE Other / P30291 WEE1 Other / Q05513 PRKCZ AGC / P34947 GRK5 AGC / O43318 MAP3K7 STE /4.5 1 identifiers used for kinases throughout the paper; 2 UniProt 1, 2 identifiers; 3 number of Bemis-Murko Frameworks (BMFs) 3 ; 4 number of actives per BMF; 5 average of Tanimoto distances per kinase data set; 6 average of IC 50 values per kinase data set; 7 hotspot value and the real maximum IC 50 value (for actives) per kinase set; 8 although NEK2 is classified by UniProt 1, 2 in the NEK group ( Human and mouse protein kinases: classification and index ; version 2015_02; see in text), here, we considered it as a member of Other according to Manning et al 4 ; 8 Atypical: PI3/PI4- kinase. Table S2. Standardization steps and parameters applied to process molecular structures using module Chemaxon Standardizer 5 Step Description Parameter 1 Remove Fragment Remove Fragment, Measure="atomCount", Method="keepLargest" 2 StripSalts 3 Dearomatize 4 Neutralize 5 Transformation Transform Isocyanate, Transform Nitroso, Transform Phosphonic,Transform Phosphonium Ylide, Transform Selenite, Transform Silicate, Transform Sulfine, Transform Sulfon, Transform Sulfonium Ylide, 6 Aromatize Type="basic" Table S3. Control parameters for random forest model generation using cforest, 6-9 in package party available in R statistical software system 10 Command cforest_control(teststat = "max", testtype = "Teststatistic", mincriterion = qnorm(0.9), ntrees = 500, mtry=15) 4
5 Table S4. Number of active and inactive compounds (according to the corresponding data source) in the training and test sets Training sets Classification test sets Virtual screening ID 1 test sets ChEMBL Actives Total Inactives 2 PubChem KinIna ChEMBL inactives ChEMBL Actives Total Inactives 3 PubChem KinIna ChEMBL Inactives PubChem KinIna
6
7 Kinase identification used throughout the article; 2 Sums of inactives from PubChemKinIna and ChEMBL inactives used for training; 3 inactives from PubChemKinIna and ChEMBL inactives used for classification testing; 4 virtual screening test sets are composed from ChEMBL actives (used in the classification test sets) and PubChemKinIna set (after removing inactives used for modeling). Table S5. Median, average and standard deviation of the number of compounds and the actives (computed per kinase set) ranked before the top 0.5% FP in the external VS test sets Cluster Num. of compounds Num. of actives retrieved Novel Actives 1 k1 278 / 302 / / 107 / / 19 / 17.5 k2 217 / 225 / / 28 / / 5 / 4.8 k3 219 / 224 / / 27 / / 5 / 3.8 k4 206 / 213 / / 17 / / 2 / 1.7 All 232 / 259 / / 64 / / 12 / Novel in the sense that these actives share BMFs which were not used in training Table S6. Wilcoxon right shift p-values (α = 0.05): row elements contain significantly higher values compared to column elements; median values are reported in parentheses Internal Classification Acc k1 k2 k3 k4 k1 (0.8725) k2 (0.8308) k3 (0.9167) k4 (0.8308) External Classification test set Acc k1 k2 k3 k4 k1 (0.8919) k2 (0.8676) k3 (0.9167) k4 (0.8919)
8 Sp k1 k2 k3 k4 k1 (0.9200) k2 (0.9038) k3 (0.9487) k4 ( ) Se k1 k2 k3 k4 k1 (0.8571) k2 (0.8421) k3 (0.9167) k4 (0.9125) AUC k1 k2 k3 k4 k1 (0.960) k2 (0.946) k3 (0.969) k4 (0.950) External Virtual Screening test set eroce k1 k2 k3 k4 k1 (0.9127) k2 (0.8900) k3 (0.9585) k4 (0.9639) TPR at 0.5% FP k1 k2 k3 k4 k1 (0.8435) k2 (0.8000) k3 (0.9375) k4 (0.9250) Table S7. Classification results of the kinase model prediction for kinase sets with actives and inactives defined as pki 7 and pki< 5, respectively, in Metz et al 11 profiling results Uniprot ID 1 Se Sp Acc AUC SD Actives Inactives P P P P O O P P P P
9 O Q96GD P P P P P P P Q P Q Q P P P Q5S Q86V P P Q Q P P Q Q9HAZ P P P Q9UM Q P Q Q Q9Y Q9Y Q9UHD Q Q Q Q9H4B Q
10 Table S8. Classification results of the kinase model prediction for kinase sets with actives and inactives defined as scaled Activity % 0.1 and > 0.9, respectively, in Anastassiadis et al 12 profiling kinase sets Uniprot ID 1 Se Sp Acc AUC SD Actives Inactives P P P24941cA Q96GD P P P Q00535p Q00535p Q
11 Figure S1. Density distribution plots of the activity values (IC50 in µm) and hot spot cutoff (red line) for the first three kinases (according to the ID number assigned in the paper) in cluster k1 (a), k2 (b), k3 (c) and k4 (d). The plot title contains the UniProt ID of the kinase and the ID number (in parenthesis) used in the paper. 11
12 Figure S2. Dendrograms resulted from Ward hierarchical clustering of 107 kinase sets in a first run (left), resulting in k4 (magenta) and a larger cluster of 103 kinase sets. These were clustered separately into 3 more groups (right) k2 (blue), k3 (red) and k1 (green). Figure S3. Accuracy (Acc), sensitivity (Se) and specificity (Sp) values of the internal (training sets) evaluation of the 107 kinase models. 12
13 Figure S4. Phylogenetic tree of the human kinases (generated using Kinome Render 13, 14 ) Target IDs and abbreviations of the kinases modeled in the current study are colored in red; two Atypical:PI3/PI4-kinase kinases, i.e., PIK3CA and PIK3CG could not be mapped and are not represented. Illustration reproduced courtesy of Cell Signaling Technology, Inc. ( 13
14 Figure S5. The distributions of (the kinase-set values) eroce (a, performed on the VS test sets) and AUC (b, performed on the classification test sets) according to the nine kinase groups. Figure S6. Venn plot (created using the online software from Microarray Center CRP-Sante) 15 of the chemical scaffolds (BMFs) contained in the PubChemKinIna (only inactives), ChEMBL (only kinase inhibitors) and ColBioS-FlavRC 16 datasets. 14
15 REFERENCES (1) UniProt, C., Reorganizing the Protein Space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012, 40, D (2) UniProtKB. (accessed Feb 4, 2015). (3) Bemis, G. W.; Murcko, M. A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, (4) Manning, G.; Whyte, D. B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, (5) ChemAxon JChem API package, version ; ChemAxon ( (6) Strobl, C.; Hothorn, T.; Zeileis, A. Party on! - A New, Conditional Variable Importance Measure for Random Forests Available in the party Package. The R Journal 2009, 1, (7) Strobl, C.; Malley, J.; Tutz, G. An Introduction to Recursive Partitioning: Rationale, Application, and Characteristics of Classification and Regression Trees, Bagging, and Random Forests. Psychol. Methods 2009, 14, (8) Strobl, C.; Boulesteix, A. L.; Zeileis, A.; Hothorn, T. Bias in Random Forest Variable Importance Measures: Illustrations, Sources and a Solution. BMC Bioinf. 2007, 8, 25. (9) Hothorn, T.; Buhlmann, P.; Dudoit, S.; Molinaro, A.; van der Laan, M. J. Survival Ensembles. Biostatistics 2006, 7, (10) R Development Core Team. R: A Language and Environment for Statistical Computing, version 3.1.1; The R Foundation for Statistical Computing: Vienna, Austria, 2014; (11) Metz, J. T.; Johnson, E. F.; Soni, N. B.; Merta, P. J.; Kifle, L.; Hajduk, P. J. Navigating the Kinome. Nat. Chem. Biol. 2011, 7,
16 (12) Anastassiadis, T.; Deacon, S. W.; Devarajan, K.; Ma, H.; Peterson, J. R. Comprehensive Assay of Kinase Catalytic Activity Reveals Features of Kinase Inhibitor Selectivity. Nat. Biotechnol. 2011, 29, (13) Chartier, M.; Chenard, T.; Barker, J.; Najmanovich, R. Kinome Render: a Stand-Alone and web- Accessible Tool to Annotate the Human Protein Kinome Tree. Peer J. 2013, 1, e126. (14) Najmanovich, R. Kinome Render [Online], (accessed Sept 28, 2015). (15) Microarray Center CRP-Sante [Online], (accessed January 2016). (16) Avram, S. I.; Pacureanu, L. M.; Bora, A.; Crisan, L.; Avram, S.; Kurunczi, L. ColBioS-FlavRC: A Collection of Bioselective Flavonoids and Related Compounds Filtered from High-Throughput Screening Outcomes. J. Chem. Inf. Model. 2014, 54,
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Supporting Information. Discovery of MK-8033: A Specific c-met/ron Dual Kinase Inhibitor with Preferential Affinity for the Activated State of c-met.
 S 1 1 Supporting Information Discovery of MK-8033: A Specific c-met/ron Dual Inhibitor with Preferential Affinity for the Activated State of c-met. Alan B. Northrup, a* Matthew H. Katcher, a Michael D.
S 1 1 Supporting Information Discovery of MK-8033: A Specific c-met/ron Dual Inhibitor with Preferential Affinity for the Activated State of c-met. Alan B. Northrup, a* Matthew H. Katcher, a Michael D.
Supporting Information. Property focused structure-based optimization of small molecule inhibitors of the proteinprotein
 Supporting Information Property focused structure-based optimization of small molecule inhibitors of the proteinprotein interaction between menin and Mixed Lineage Leukemia (MLL) Dmitry Borkin a, Jonathan
Supporting Information Property focused structure-based optimization of small molecule inhibitors of the proteinprotein interaction between menin and Mixed Lineage Leukemia (MLL) Dmitry Borkin a, Jonathan
Supporting Information
 Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Bayesian statistics. DS GA 1002 Probability and Statistics for Data Science.
 Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Bayesian statistics DS GA 1002 Probability and Statistics for Data Science http://www.cims.nyu.edu/~cfgranda/pages/dsga1002_fall17 Carlos Fernandez-Granda Frequentist vs Bayesian statistics In frequentist
Supplementary Appendix
 Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Supplementary Appendix Measuring crisis risk using conditional copulas: An empirical analysis of the 2008 shipping crisis Sebastian Opitz, Henry Seidel and Alexander Szimayer Model specification Table
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) Cyclopentadienyl iron dicarbonyl (CpFe(CO) 2 ) derivatives
Chapter 1 Introduction to Observational Studies Part 2 Cross-Sectional Selection Bias Adjustment
 Contents Preface ix Part 1 Introduction Chapter 1 Introduction to Observational Studies... 3 1.1 Observational vs. Experimental Studies... 3 1.2 Issues in Observational Studies... 5 1.3 Study Design...
Contents Preface ix Part 1 Introduction Chapter 1 Introduction to Observational Studies... 3 1.1 Observational vs. Experimental Studies... 3 1.2 Issues in Observational Studies... 5 1.3 Study Design...
EPL 603 TOPICS IN SOFTWARE ENGINEERING. Lab 5: Component Adaptation Environment (COPE)
 EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
EPL 603 TOPICS IN SOFTWARE ENGINEERING Lab 5: Component Adaptation Environment (COPE) Performing Static Analysis 1 Class Name: The fully qualified name of the specific class Type: The type of the class
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ
 ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΤΜΗΜΑ ΑΓΡΟΤΙΚΗΣ ΟΙΚΟΝΟΜΙΑΣ & ΑΝΑΠΤΥΞΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΟΛΟΚΛΗΡΩΜΕΝΗ ΑΝΑΠΤΥΞΗ & ΔΙΑΧΕΙΡΙΣΗ ΤΟΥ ΑΓΡΟΤΙΚΟΥ ΧΩΡΟΥ» ΜΕΤΑΠΤΥΧΙΑΚΗ ΕΡΓΑΣΙΑ «Οικονομετρική διερεύνηση
Statistics 104: Quantitative Methods for Economics Formula and Theorem Review
 Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
Harvard College Statistics 104: Quantitative Methods for Economics Formula and Theorem Review Tommy MacWilliam, 13 tmacwilliam@college.harvard.edu March 10, 2011 Contents 1 Introduction to Data 5 1.1 Sample
VBA Microsoft Excel. J. Comput. Chem. Jpn., Vol. 5, No. 1, pp (2006)
 J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
J. Comput. Chem. Jpn., Vol. 5, No. 1, pp. 29 38 (2006) Microsoft Excel, 184-8588 2-24-16 e-mail: yosimura@cc.tuat.ac.jp (Received: July 28, 2005; Accepted for publication: October 24, 2005; Published on
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
SCITECH Volume 13, Issue 2 RESEARCH ORGANISATION Published online: March 29, 2018
 Journal of rogressive Research in Mathematics(JRM) ISSN: 2395-028 SCITECH Volume 3, Issue 2 RESEARCH ORGANISATION ublished online: March 29, 208 Journal of rogressive Research in Mathematics www.scitecresearch.com/journals
Journal of rogressive Research in Mathematics(JRM) ISSN: 2395-028 SCITECH Volume 3, Issue 2 RESEARCH ORGANISATION ublished online: March 29, 208 Journal of rogressive Research in Mathematics www.scitecresearch.com/journals
Statistical Inference I Locally most powerful tests
 Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
Statistical Inference I Locally most powerful tests Shirsendu Mukherjee Department of Statistics, Asutosh College, Kolkata, India. shirsendu st@yahoo.co.in So far we have treated the testing of one-sided
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ
 ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ Γηπισκαηηθή Δξγαζία ηνπ Φνηηεηή ηνπ ηκήκαηνο Ζιεθηξνιόγσλ Μεραληθώλ θαη Σερλνινγίαο Ζιεθηξνληθώλ
ΠΑΝΔΠΗΣΖΜΗΟ ΠΑΣΡΩΝ ΣΜΖΜΑ ΖΛΔΚΣΡΟΛΟΓΩΝ ΜΖΥΑΝΗΚΩΝ ΚΑΗ ΣΔΥΝΟΛΟΓΗΑ ΤΠΟΛΟΓΗΣΩΝ ΣΟΜΔΑ ΤΣΖΜΑΣΩΝ ΖΛΔΚΣΡΗΚΖ ΔΝΔΡΓΔΗΑ Γηπισκαηηθή Δξγαζία ηνπ Φνηηεηή ηνπ ηκήκαηνο Ζιεθηξνιόγσλ Μεραληθώλ θαη Σερλνινγίαο Ζιεθηξνληθώλ
Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo
 Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
Bull. Earthq. Res. Inst. Univ. Tokyo Vol. 2.,**3 pp.,,3,.* * +, -. +, -. Resurvey of Possible Seismic Fissures in the Old-Edo River in Tokyo Kunihiko Shimazaki *, Tsuyoshi Haraguchi, Takeo Ishibe +, -.
[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,
![[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13, [11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,](/thumbs/89/98694254.jpg) in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ
 ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
ΤΕΧΝΟΛΟΓΙΚΟ ΕΚΠΑΙ ΕΥΤΙΚΟ Ι ΡΥΜΑ ΚΡΗΤΗΣ ΤΜΗΜΑ ΦΥΣΙΚΩΝ ΠΟΡΩΝ ΚΑΙ ΠΕΡΙΒΑΛΛΟΝΤΟΣ ΕΦΑΡΜΟΓΗ ΕΥΤΕΡΟΒΑΘΜΙΑ ΕΠΕΞΕΡΓΑΣΜΕΝΩΝ ΥΓΡΩΝ ΑΠΟΒΛΗΤΩΝ ΣΕ ΦΥΣΙΚΑ ΣΥΣΤΗΜΑΤΑ ΚΛΙΝΗΣ ΚΑΛΑΜΙΩΝ ΕΠΙΜΕΛΕΙΑ: ΑΡΜΕΝΑΚΑΣ ΜΑΡΙΝΟΣ ΧΑΝΙΑ
HIV HIV HIV HIV AIDS 3 :.1 /-,**1 +332
 ,**1 The Japanese Society for AIDS Research The Journal of AIDS Research +,, +,, +,, + -. / 0 1 +, -. / 0 1 : :,**- +,**. 1..+ - : +** 22 HIV AIDS HIV HIV AIDS : HIV AIDS HIV :HIV AIDS 3 :.1 /-,**1 HIV
,**1 The Japanese Society for AIDS Research The Journal of AIDS Research +,, +,, +,, + -. / 0 1 +, -. / 0 1 : :,**- +,**. 1..+ - : +** 22 HIV AIDS HIV HIV AIDS : HIV AIDS HIV :HIV AIDS 3 :.1 /-,**1 HIV
Οι απόψεις και τα συμπεράσματα που περιέχονται σε αυτό το έγγραφο, εκφράζουν τον συγγραφέα και δεν πρέπει να ερμηνευτεί ότι αντιπροσωπεύουν τις
 Οι απόψεις και τα συμπεράσματα που περιέχονται σε αυτό το έγγραφο, εκφράζουν τον συγγραφέα και δεν πρέπει να ερμηνευτεί ότι αντιπροσωπεύουν τις επίσημες θέσεις των εξεταστών. i ΠΡΟΛΟΓΟΣ ΕΥΧΑΡΙΣΤΙΕΣ Η παρούσα
Οι απόψεις και τα συμπεράσματα που περιέχονται σε αυτό το έγγραφο, εκφράζουν τον συγγραφέα και δεν πρέπει να ερμηνευτεί ότι αντιπροσωπεύουν τις επίσημες θέσεις των εξεταστών. i ΠΡΟΛΟΓΟΣ ΕΥΧΑΡΙΣΤΙΕΣ Η παρούσα
Test Data Management in Practice
 Problems, Concepts, and the Swisscom Test Data Organizer Do you have issues with your legal and compliance department because test environments contain sensitive data outsourcing partners must not see?
Problems, Concepts, and the Swisscom Test Data Organizer Do you have issues with your legal and compliance department because test environments contain sensitive data outsourcing partners must not see?
WORKING P A P E R. Learning your Child s Price. Evidence from Data on Projected Dowry in Rural India A. V. CHARI AND ANNEMIE MAERTENS WR-899
 WORKING P A P E R Learning your Child s Price Evidence from Data on Projected Dowry in Rural India A. V. CHARI AND ANNEMIE MAERTENS WR-899 November 2011 This product is part of the RAND Labor and Population
WORKING P A P E R Learning your Child s Price Evidence from Data on Projected Dowry in Rural India A. V. CHARI AND ANNEMIE MAERTENS WR-899 November 2011 This product is part of the RAND Labor and Population
An Introduction to Spatial Statistics: Data Types, Statistical Tools and Computer Software
 An Introduction to Spatial Statistics: Data Types, Statistical Tools and Computer Software Moira A. Mugglestone MRC Institute for Environment and Health mam14@le.ac.uk http://www.le.ac.uk/ieh/ spatial.1
An Introduction to Spatial Statistics: Data Types, Statistical Tools and Computer Software Moira A. Mugglestone MRC Institute for Environment and Health mam14@le.ac.uk http://www.le.ac.uk/ieh/ spatial.1
On a four-dimensional hyperbolic manifold with finite volume
 BULETINUL ACADEMIEI DE ŞTIINŢE A REPUBLICII MOLDOVA. MATEMATICA Numbers 2(72) 3(73), 2013, Pages 80 89 ISSN 1024 7696 On a four-dimensional hyperbolic manifold with finite volume I.S.Gutsul Abstract. In
BULETINUL ACADEMIEI DE ŞTIINŢE A REPUBLICII MOLDOVA. MATEMATICA Numbers 2(72) 3(73), 2013, Pages 80 89 ISSN 1024 7696 On a four-dimensional hyperbolic manifold with finite volume I.S.Gutsul Abstract. In
Partial Trace and Partial Transpose
 Partial Trace and Partial Transpose by José Luis Gómez-Muñoz http://homepage.cem.itesm.mx/lgomez/quantum/ jose.luis.gomez@itesm.mx This document is based on suggestions by Anirban Das Introduction This
Partial Trace and Partial Transpose by José Luis Gómez-Muñoz http://homepage.cem.itesm.mx/lgomez/quantum/ jose.luis.gomez@itesm.mx This document is based on suggestions by Anirban Das Introduction This
 1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
1 1 1 2 1 2 2 1 43 123 5 122 3 1 312 1 1 122 1 1 1 1 6 1 7 1 6 1 7 1 3 4 2 312 43 4 3 3 1 1 4 1 1 52 122 54 124 8 1 3 1 1 1 1 1 152 1 1 1 1 1 1 152 1 5 1 152 152 1 1 3 9 1 159 9 13 4 5 1 122 1 4 122 5
4.6 Autoregressive Moving Average Model ARMA(1,1)
 84 CHAPTER 4. STATIONARY TS MODELS 4.6 Autoregressive Moving Average Model ARMA(,) This section is an introduction to a wide class of models ARMA(p,q) which we will consider in more detail later in this
84 CHAPTER 4. STATIONARY TS MODELS 4.6 Autoregressive Moving Average Model ARMA(,) This section is an introduction to a wide class of models ARMA(p,q) which we will consider in more detail later in this
Zebra reaction or the recipe for heterodimeric zinc complexes synthesis
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 05 Supporting Information Zebra reaction or the recipe for heterodimeric zinc complexes synthesis
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 05 Supporting Information Zebra reaction or the recipe for heterodimeric zinc complexes synthesis
Study of In-vehicle Sound Field Creation by Simultaneous Equation Method
 Study of In-vehicle Sound Field Creation by Simultaneous Equation Method Kensaku FUJII Isao WAKABAYASI Tadashi UJINO Shigeki KATO Abstract FUJITSU TEN Limited has developed "TOYOTA remium Sound System"
Study of In-vehicle Sound Field Creation by Simultaneous Equation Method Kensaku FUJII Isao WAKABAYASI Tadashi UJINO Shigeki KATO Abstract FUJITSU TEN Limited has developed "TOYOTA remium Sound System"
Notes on the Open Economy
 Notes on the Open Econom Ben J. Heijdra Universit of Groningen April 24 Introduction In this note we stud the two-countr model of Table.4 in more detail. restated here for convenience. The model is Table.4.
Notes on the Open Econom Ben J. Heijdra Universit of Groningen April 24 Introduction In this note we stud the two-countr model of Table.4 in more detail. restated here for convenience. The model is Table.4.
Reminders: linear functions
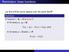 Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ BIOCHEMICAL JOURNAL
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Description of the PX-HC algorithm
 A Description of the PX-HC algorithm Let N = C c= N c and write C Nc K c= i= k= as, the Gibbs sampling algorithm at iteration m for continuous outcomes: Step A: For =,, J, draw θ m in the following steps:
A Description of the PX-HC algorithm Let N = C c= N c and write C Nc K c= i= k= as, the Gibbs sampling algorithm at iteration m for continuous outcomes: Step A: For =,, J, draw θ m in the following steps:
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models
 Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Longitudinal Changes in Component Processes of Working Memory
 This Accepted Manuscript has not been copyedited and formatted. The final version may differ from this version. Research Article: New Research Cognition and Behavior Longitudinal Changes in Component Processes
This Accepted Manuscript has not been copyedited and formatted. The final version may differ from this version. Research Article: New Research Cognition and Behavior Longitudinal Changes in Component Processes
5.4 The Poisson Distribution.
 The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
The worst thing you can do about a situation is nothing. Sr. O Shea Jackson 5.4 The Poisson Distribution. Description of the Poisson Distribution Discrete probability distribution. The random variable
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1
 Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
Main source: "Discrete-time systems and computer control" by Α. ΣΚΟΔΡΑΣ ΨΗΦΙΑΚΟΣ ΕΛΕΓΧΟΣ ΔΙΑΛΕΞΗ 4 ΔΙΑΦΑΝΕΙΑ 1 A Brief History of Sampling Research 1915 - Edmund Taylor Whittaker (1873-1956) devised a
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
BMI/CS 776 Lecture #14: Multiple Alignment - MUSCLE. Colin Dewey
 BMI/CS 776 Lecture #14: Multiple Alignment - MUSCLE Colin Dewey 2007.03.08 1 Importance of protein multiple alignment Phylogenetic tree estimation Prediction of protein secondary structure Critical residue
BMI/CS 776 Lecture #14: Multiple Alignment - MUSCLE Colin Dewey 2007.03.08 1 Importance of protein multiple alignment Phylogenetic tree estimation Prediction of protein secondary structure Critical residue
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΤΕΧΝΟΛΟΓΙΑΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΥΠΟΛΟΓΙΣΤΩΝ Σημασιολογική Συσταδοποίηση Αντικειμένων Με Χρήση Οντολογικών Περιγραφών.
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΤΕΧΝΟΛΟΓΙΑΣ ΠΛΗΡΟΦΟΡΙΚΗΣ ΚΑΙ ΥΠΟΛΟΓΙΣΤΩΝ Σημασιολογική Συσταδοποίηση Αντικειμένων Με Χρήση Οντολογικών Περιγραφών.
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Η Διαγενειακή Αλληλεπίδραση Τρίτης Γενιάς και Τρίτης Ηλικίας και οι Αντοχές της Ελληνοαυστραλιανής Ταυτότητας
 Kyriakopoulou-Baltatzi, C., 2013. Η Διαγενειακή Αλληλεπίδραση Τρίτης Γενιάς και Τρίτης Ηλικίας και οι Αντοχές της Ελληνοαυστραλιανής Ταυτότητας. Journal of Modern Greek Studies - Special Issue, 346-361.
Kyriakopoulou-Baltatzi, C., 2013. Η Διαγενειακή Αλληλεπίδραση Τρίτης Γενιάς και Τρίτης Ηλικίας και οι Αντοχές της Ελληνοαυστραλιανής Ταυτότητας. Journal of Modern Greek Studies - Special Issue, 346-361.
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp.
 SUPPLEMENTARY MATERIAL New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp. Mohamed-Elamir F. Hegazy a,*, Amira M. Gamal-Eldeen b, Tarik A. Mohamed a, Montaser A. Alhammady c, Abuzeid A.
SUPPLEMENTARY MATERIAL New Cytotoxic Constituents from the Red Sea Soft Coral Nephthea sp. Mohamed-Elamir F. Hegazy a,*, Amira M. Gamal-Eldeen b, Tarik A. Mohamed a, Montaser A. Alhammady c, Abuzeid A.
Schedulability Analysis Algorithm for Timing Constraint Workflow Models
 CIMS Vol.8No.72002pp.527-532 ( 100084) Petri Petri F270.7 A Schedulability Analysis Algorithm for Timing Constraint Workflow Models Li Huifang and Fan Yushun (Department of Automation, Tsinghua University,
CIMS Vol.8No.72002pp.527-532 ( 100084) Petri Petri F270.7 A Schedulability Analysis Algorithm for Timing Constraint Workflow Models Li Huifang and Fan Yushun (Department of Automation, Tsinghua University,
MSM Men who have Sex with Men HIV -
 ,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
,**, The Japanese Society for AIDS Research The Journal of AIDS Research HIV,0 + + + + +,,, +, : HIV : +322,*** HIV,0,, :., n,0,,. + 2 2, CD. +3-ml n,, AIDS 3 ARC 3 +* 1. A, MSM Men who have Sex with Men
derivation of the Laplacian from rectangular to spherical coordinates
 derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
derivation of the Laplacian from rectangular to spherical coordinates swapnizzle 03-03- :5:43 We begin by recognizing the familiar conversion from rectangular to spherical coordinates (note that φ is used
ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE ΑΠΟ ΑΤΟΜΑ ΜΕ ΤΥΦΛΩΣΗ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
ΠΑΝΕΠΙΣΤΗΜΙΟ ΜΑΚΕΔΟΝΙΑΣ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΕΚΠΑΙΔΕΥΤΙΚΗΣ ΚΑΙ ΚΟΙΝΩΝΙΚΗΣ ΠΟΛΙΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΠΑΡΑΜΕΤΡΟΙ ΕΠΗΡΕΑΣΜΟΥ ΤΗΣ ΑΝΑΓΝΩΣΗΣ- ΑΠΟΚΩΔΙΚΟΠΟΙΗΣΗΣ ΤΗΣ BRAILLE
Structure-Metabolism-Relationships in the microsomal clearance of. piperazin-1-ylpyridazines
 Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2017 Structure-Metabolism-Relationships in the microsomal clearance of piperazin-1-ylpyridazines
Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2017 Structure-Metabolism-Relationships in the microsomal clearance of piperazin-1-ylpyridazines
Ανάκτηση Πληροφορίας
 Ανάκτηση Πληροφορίας Αποτίμηση Αποτελεσματικότητας Μέτρα Απόδοσης Precision = # σχετικών κειμένων που επιστρέφονται # κειμένων που επιστρέφονται Recall = # σχετικών κειμένων που επιστρέφονται # συνολικών
Ανάκτηση Πληροφορίας Αποτίμηση Αποτελεσματικότητας Μέτρα Απόδοσης Precision = # σχετικών κειμένων που επιστρέφονται # κειμένων που επιστρέφονται Recall = # σχετικών κειμένων που επιστρέφονται # συνολικών
Solution Series 9. i=1 x i and i=1 x i.
 Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
Lecturer: Prof. Dr. Mete SONER Coordinator: Yilin WANG Solution Series 9 Q1. Let α, β >, the p.d.f. of a beta distribution with parameters α and β is { Γ(α+β) Γ(α)Γ(β) f(x α, β) xα 1 (1 x) β 1 for < x
Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων. Εξάμηνο 7 ο
 Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων Εξάμηνο 7 ο Procedures and Functions Stored procedures and functions are named blocks of code that enable you to group and organize a series of SQL and PL/SQL
Εργαστήριο Ανάπτυξης Εφαρμογών Βάσεων Δεδομένων Εξάμηνο 7 ο Procedures and Functions Stored procedures and functions are named blocks of code that enable you to group and organize a series of SQL and PL/SQL
Summary of the model specified
 Program: HLM 7 Hierarchical Linear and Nonlinear Modeling Authors: Stephen Raudenbush, Tony Bryk, & Richard Congdon Publisher: Scientific Software International, Inc. (c) 2010 techsupport@ssicentral.com
Program: HLM 7 Hierarchical Linear and Nonlinear Modeling Authors: Stephen Raudenbush, Tony Bryk, & Richard Congdon Publisher: Scientific Software International, Inc. (c) 2010 techsupport@ssicentral.com
Optimizing Microwave-assisted Extraction Process for Paprika Red Pigments Using Response Surface Methodology
 2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
2012 34 2 382-387 http / /xuebao. jxau. edu. cn Acta Agriculturae Universitatis Jiangxiensis E - mail ndxb7775@ sina. com 212018 105 W 42 2 min 0. 631 TS202. 3 A 1000-2286 2012 02-0382 - 06 Optimizing
 CSAE WPS/2009-06 Figure 1: Cut Flower Exports from Kenya, 1995-2007 Table 1: Firms in Areas with and w/out Conflict Panel A - Export Records Variable Observations Mean in No-Conflict
CSAE WPS/2009-06 Figure 1: Cut Flower Exports from Kenya, 1995-2007 Table 1: Firms in Areas with and w/out Conflict Panel A - Export Records Variable Observations Mean in No-Conflict
Electrolyzed-Reduced Water as Artificial Hot Spring Water
 /-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
/-,**- + +/ 0 +, - + + +, - + +. ++,3 +/. +. Electrolyzed-Reduced Water as Artificial Hot Spring Water Shoichi OKOUCHI +, Daisuke TAKEZAKI +, Hideyuki OHNAMI +, Yuhkoh AGISHI,, Yasuo KANROJI -, and Shigeo
HW 3 Solutions 1. a) I use the auto.arima R function to search over models using AIC and decide on an ARMA(3,1)
 HW 3 Solutions a) I use the autoarima R function to search over models using AIC and decide on an ARMA3,) b) I compare the ARMA3,) to ARMA,0) ARMA3,) does better in all three criteria c) The plot of the
HW 3 Solutions a) I use the autoarima R function to search over models using AIC and decide on an ARMA3,) b) I compare the ARMA3,) to ARMA,0) ARMA3,) does better in all three criteria c) The plot of the
Vol. 31,No JOURNAL OF CHINA UNIVERSITY OF SCIENCE AND TECHNOLOGY Feb
 Ξ 31 Vol 31,No 1 2 0 0 1 2 JOURNAL OF CHINA UNIVERSITY OF SCIENCE AND TECHNOLOGY Feb 2 0 0 1 :025322778 (2001) 0120016205 (, 230026) : Q ( m 1, m 2,, m n ) k = m 1 + m 2 + + m n - n : Q ( m 1, m 2,, m
Ξ 31 Vol 31,No 1 2 0 0 1 2 JOURNAL OF CHINA UNIVERSITY OF SCIENCE AND TECHNOLOGY Feb 2 0 0 1 :025322778 (2001) 0120016205 (, 230026) : Q ( m 1, m 2,, m n ) k = m 1 + m 2 + + m n - n : Q ( m 1, m 2,, m
Supporting Information
 Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
Supporting Information rigin of the Regio- and Stereoselectivity of Allylic Substitution of rganocopper Reagents Naohiko Yoshikai, Song-Lin Zhang, and Eiichi Nakamura* Department of Chemistry, The University
Homomorphism in Intuitionistic Fuzzy Automata
 International Journal of Fuzzy Mathematics Systems. ISSN 2248-9940 Volume 3, Number 1 (2013), pp. 39-45 Research India Publications http://www.ripublication.com/ijfms.htm Homomorphism in Intuitionistic
International Journal of Fuzzy Mathematics Systems. ISSN 2248-9940 Volume 3, Number 1 (2013), pp. 39-45 Research India Publications http://www.ripublication.com/ijfms.htm Homomorphism in Intuitionistic
«ΑΓΡΟΤΟΥΡΙΣΜΟΣ ΚΑΙ ΤΟΠΙΚΗ ΑΝΑΠΤΥΞΗ: Ο ΡΟΛΟΣ ΤΩΝ ΝΕΩΝ ΤΕΧΝΟΛΟΓΙΩΝ ΣΤΗΝ ΠΡΟΩΘΗΣΗ ΤΩΝ ΓΥΝΑΙΚΕΙΩΝ ΣΥΝΕΤΑΙΡΙΣΜΩΝ»
 I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
I ΑΡΙΣΤΟΤΕΛΕΙΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΟΝΙΚΗΣ ΣΧΟΛΗ ΝΟΜΙΚΩΝ ΟΙΚΟΝΟΜΙΚΩΝ ΚΑΙ ΠΟΛΙΤΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΤΜΗΜΑ ΟΙΚΟΝΟΜΙΚΩΝ ΕΠΙΣΤΗΜΩΝ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΗΝ «ΔΙΟΙΚΗΣΗ ΚΑΙ ΟΙΚΟΝΟΜΙΑ» ΚΑΤΕΥΘΥΝΣΗ: ΟΙΚΟΝΟΜΙΚΗ
2 Composition. Invertible Mappings
 Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Arkansas Tech University MATH 4033: Elementary Modern Algebra Dr. Marcel B. Finan Composition. Invertible Mappings In this section we discuss two procedures for creating new mappings from old ones, namely,
Research on Economics and Management
 36 5 2015 5 Research on Economics and Management Vol. 36 No. 5 May 2015 490 490 F323. 9 A DOI:10.13502/j.cnki.issn1000-7636.2015.05.007 1000-7636 2015 05-0052 - 10 2008 836 70% 1. 2 2010 1 2 3 2015-03
36 5 2015 5 Research on Economics and Management Vol. 36 No. 5 May 2015 490 490 F323. 9 A DOI:10.13502/j.cnki.issn1000-7636.2015.05.007 1000-7636 2015 05-0052 - 10 2008 836 70% 1. 2 2010 1 2 3 2015-03
Biostatistics for Health Sciences Review Sheet
 Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
Biostatistics for Health Sciences Review Sheet http://mathvault.ca June 1, 2017 Contents 1 Descriptive Statistics 2 1.1 Variables.............................................. 2 1.1.1 Qualitative........................................
ΜΕΛΕΤΗ ΤΗΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΣΥΝΤΑΓΟΓΡΑΦΗΣΗΣ ΚΑΙ Η ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΕΦΑΡΜΟΓΗΣ ΤΗΣ ΣΤΗΝ ΕΛΛΑΔΑ: Ο.Α.Ε.Ε. ΠΕΡΙΦΕΡΕΙΑ ΠΕΛΟΠΟΝΝΗΣΟΥ ΚΑΣΚΑΦΕΤΟΥ ΣΩΤΗΡΙΑ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΩΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΔΙΟΙΚΗΣΗ ΤΗΣ ΥΓΕΙΑΣ ΤΕΙ ΠΕΙΡΑΙΑ ΜΕΛΕΤΗ ΤΗΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΣΥΝΤΑΓΟΓΡΑΦΗΣΗΣ ΚΑΙ Η ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΕΦΑΡΜΟΓΗΣ ΤΗΣ ΣΤΗΝ ΕΛΛΑΔΑ: Ο.Α.Ε.Ε. ΠΕΡΙΦΕΡΕΙΑ ΠΕΛΟΠΟΝΝΗΣΟΥ
ΠΑΝΕΠΙΣΤΗΜΙΟ ΠΕΙΡΑΙΩΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΔΙΟΙΚΗΣΗ ΤΗΣ ΥΓΕΙΑΣ ΤΕΙ ΠΕΙΡΑΙΑ ΜΕΛΕΤΗ ΤΗΣ ΗΛΕΚΤΡΟΝΙΚΗΣ ΣΥΝΤΑΓΟΓΡΑΦΗΣΗΣ ΚΑΙ Η ΔΙΕΡΕΥΝΗΣΗ ΤΗΣ ΕΦΑΡΜΟΓΗΣ ΤΗΣ ΣΤΗΝ ΕΛΛΑΔΑ: Ο.Α.Ε.Ε. ΠΕΡΙΦΕΡΕΙΑ ΠΕΛΟΠΟΝΝΗΣΟΥ
A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads. Electronic Supplementary Material
 A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads Electronic Supplementary Material Ivan Jarić 1,2*, Jörn Gessner 1 and Mirjana Lenhardt 3 1 Leibniz-Institute
A life-table metamodel to support management of data deficient species, exemplified in sturgeons and shads Electronic Supplementary Material Ivan Jarić 1,2*, Jörn Gessner 1 and Mirjana Lenhardt 3 1 Leibniz-Institute
ES440/ES911: CFD. Chapter 5. Solution of Linear Equation Systems
 ES440/ES911: CFD Chapter 5. Solution of Linear Equation Systems Dr Yongmann M. Chung http://www.eng.warwick.ac.uk/staff/ymc/es440.html Y.M.Chung@warwick.ac.uk School of Engineering & Centre for Scientific
ES440/ES911: CFD Chapter 5. Solution of Linear Equation Systems Dr Yongmann M. Chung http://www.eng.warwick.ac.uk/staff/ymc/es440.html Y.M.Chung@warwick.ac.uk School of Engineering & Centre for Scientific
Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic acids with the assistance of isocyanide
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Copper-catalyzed formal O-H insertion reaction of α-diazo-1,3-dicarb- onyl compounds to carboxylic
The Simply Typed Lambda Calculus
 Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
Type Inference Instead of writing type annotations, can we use an algorithm to infer what the type annotations should be? That depends on the type system. For simple type systems the answer is yes, and
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Instruction Execution Times
 1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
1 C Execution Times InThisAppendix... Introduction DL330 Execution Times DL330P Execution Times DL340 Execution Times C-2 Execution Times Introduction Data Registers This appendix contains several tables
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ
 ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
ΕΙΣΑΓΩΓΗ ΣΤΗ ΣΤΑΤΙΣΤΙΚΗ ΑΝΑΛΥΣΗ ΕΛΕΝΑ ΦΛΟΚΑ Επίκουρος Καθηγήτρια Τµήµα Φυσικής, Τοµέας Φυσικής Περιβάλλοντος- Μετεωρολογίας ΓΕΝΙΚΟΙ ΟΡΙΣΜΟΙ Πληθυσµός Σύνολο ατόµων ή αντικειµένων στα οποία αναφέρονται
EE101: Resonance in RLC circuits
 EE11: Resonance in RLC circuits M. B. Patil mbatil@ee.iitb.ac.in www.ee.iitb.ac.in/~sequel Deartment of Electrical Engineering Indian Institute of Technology Bombay I V R V L V C I = I m = R + jωl + 1/jωC
EE11: Resonance in RLC circuits M. B. Patil mbatil@ee.iitb.ac.in www.ee.iitb.ac.in/~sequel Deartment of Electrical Engineering Indian Institute of Technology Bombay I V R V L V C I = I m = R + jωl + 1/jωC
difluoroboranyls derived from amides carrying donor group Supporting Information
 The influence of the π-conjugated spacer on photophysical properties of difluoroboranyls derived from amides carrying donor group Supporting Information Anna Maria Grabarz a Adèle D. Laurent b, Beata Jędrzejewska
The influence of the π-conjugated spacer on photophysical properties of difluoroboranyls derived from amides carrying donor group Supporting Information Anna Maria Grabarz a Adèle D. Laurent b, Beata Jędrzejewska
 22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
22 .5 Real consumption.5 Real residential investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.5 Real house prices.5 Real fixed investment.5.5.5 965 975 985 995 25.5 965 975 985 995 25.3 Inflation
6.1. Dirac Equation. Hamiltonian. Dirac Eq.
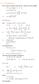 6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
6.1. Dirac Equation Ref: M.Kaku, Quantum Field Theory, Oxford Univ Press (1993) η μν = η μν = diag(1, -1, -1, -1) p 0 = p 0 p = p i = -p i p μ p μ = p 0 p 0 + p i p i = E c 2 - p 2 = (m c) 2 H = c p 2
EM Baum-Welch. Step by Step the Baum-Welch Algorithm and its Application 2. HMM Baum-Welch. Baum-Welch. Baum-Welch Baum-Welch.
 Baum-Welch Step by Step the Baum-Welch Algorithm and its Application Jin ichi MURAKAMI EM EM EM Baum-Welch Baum-Welch Baum-Welch Baum-Welch, EM 1. EM 2. HMM EM (Expectationmaximization algorithm) 1 3.
Baum-Welch Step by Step the Baum-Welch Algorithm and its Application Jin ichi MURAKAMI EM EM EM Baum-Welch Baum-Welch Baum-Welch Baum-Welch, EM 1. EM 2. HMM EM (Expectationmaximization algorithm) 1 3.
A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 A facile and general route to 3-((trifluoromethyl)thio)benzofurans and 3-((trifluoromethyl)thio)benzothiophenes
 n n 1 n+1 2 2 Farmers in Random Insurance Group Farmers in Random Insurance Group Insured Plots Control Plots 1st Choice Plots Insured plot Adverse Selection Moral Hazard Control plot 1st Choice Plots
n n 1 n+1 2 2 Farmers in Random Insurance Group Farmers in Random Insurance Group Insured Plots Control Plots 1st Choice Plots Insured plot Adverse Selection Moral Hazard Control plot 1st Choice Plots
Si + Al Mg Fe + Mn +Ni Ca rim Ca p.f.u
 .6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
.6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
ΣΔΥΝΟΛΟΓΗΚΟ ΔΚΠΑΗΓΔΤΣΗΚΟ ΗΓΡΤΜΑ ΗΟΝΗΧΝ ΝΖΧΝ «ΗΣΟΔΛΗΓΔ ΠΟΛΗΣΗΚΖ ΔΠΗΚΟΗΝΧΝΗΑ:ΜΔΛΔΣΖ ΚΑΣΑΚΔΤΖ ΔΡΓΑΛΔΗΟΤ ΑΞΗΟΛΟΓΖΖ» ΠΣΤΥΗΑΚΖ ΔΡΓΑΗΑ ΔΤΑΓΓΔΛΗΑ ΣΔΓΟΤ
 [Type the abstract of the document here. The abstract is typically a short summary of the contents of the document. Type the abstract of the document here. The abstract is typically a short summary of
[Type the abstract of the document here. The abstract is typically a short summary of the contents of the document. Type the abstract of the document here. The abstract is typically a short summary of
HOMEWORK 4 = G. In order to plot the stress versus the stretch we define a normalized stretch:
 HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
HOMEWORK 4 Problem a For the fast loading case, we want to derive the relationship between P zz and λ z. We know that the nominal stress is expressed as: P zz = ψ λ z where λ z = λ λ z. Therefore, applying
Toward the Quantitative Study of Hydrothermal Systems An Approach to Understand Hydrothermal Systems
 J. Hot Spring Sci. 0*,0+,1+,*+* + Toward the Quantitative Study of Hydrothermal Systems An Approach to Understand Hydrothermal Systems Sachio E + HARA Abstract The quantitative understanding of hydrothermal
J. Hot Spring Sci. 0*,0+,1+,*+* + Toward the Quantitative Study of Hydrothermal Systems An Approach to Understand Hydrothermal Systems Sachio E + HARA Abstract The quantitative understanding of hydrothermal
Medium Data on Big Data
 IT 17081 Examensarbete 15 hp November 2017 Medium Data on Big Data Predicting Disk Failures in CERNs NetApp-based Data Storage System Albin Stjerna Institutionen för informationsteknologi Department of
IT 17081 Examensarbete 15 hp November 2017 Medium Data on Big Data Predicting Disk Failures in CERNs NetApp-based Data Storage System Albin Stjerna Institutionen för informationsteknologi Department of
Aquinas College. Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET
 Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Ελαφρές κυψελωτές πλάκες - ένα νέο προϊόν για την επιπλοποιία και ξυλουργική. ΒΑΣΙΛΕΙΟΥ ΒΑΣΙΛΕΙΟΣ και ΜΠΑΡΜΠΟΥΤΗΣ ΙΩΑΝΝΗΣ
 Ελαφρές κυψελωτές πλάκες - ένα νέο προϊόν για την επιπλοποιία και ξυλουργική ΒΑΣΙΛΕΙΟΥ ΒΑΣΙΛΕΙΟΣ και ΜΠΑΡΜΠΟΥΤΗΣ ΙΩΑΝΝΗΣ Αριστοτέλειο Πανεπιστήµιο Θεσσαλονίκης Σχολή ασολογίας και Φυσικού Περιβάλλοντος,
Ελαφρές κυψελωτές πλάκες - ένα νέο προϊόν για την επιπλοποιία και ξυλουργική ΒΑΣΙΛΕΙΟΥ ΒΑΣΙΛΕΙΟΣ και ΜΠΑΡΜΠΟΥΤΗΣ ΙΩΑΝΝΗΣ Αριστοτέλειο Πανεπιστήµιο Θεσσαλονίκης Σχολή ασολογίας και Φυσικού Περιβάλλοντος,
Biodiesel quality and EN 14214:2012
 3η Ενότητα: «Αγορά Βιοκαυσίμων στην Ελλάδα: Τάσεις και Προοπτικές» Biodiesel quality and EN 14214:2012 Dr. Hendrik Stein Pilot Plant Manager, ASG Analytik Content Introduction Development of the Biodiesel
3η Ενότητα: «Αγορά Βιοκαυσίμων στην Ελλάδα: Τάσεις και Προοπτικές» Biodiesel quality and EN 14214:2012 Dr. Hendrik Stein Pilot Plant Manager, ASG Analytik Content Introduction Development of the Biodiesel
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS
 CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
CHAPTER 48 APPLICATIONS OF MATRICES AND DETERMINANTS EXERCISE 01 Page 545 1. Use matrices to solve: 3x + 4y x + 5y + 7 3x + 4y x + 5y 7 Hence, 3 4 x 0 5 y 7 The inverse of 3 4 5 is: 1 5 4 1 5 4 15 8 3
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Buried Markov Model Pairwise
 Buried Markov Model 1 2 2 HMM Buried Markov Model J. Bilmes Buried Markov Model Pairwise 0.6 0.6 1.3 Structuring Model for Speech Recognition using Buried Markov Model Takayuki Yamamoto, 1 Tetsuya Takiguchi
Buried Markov Model 1 2 2 HMM Buried Markov Model J. Bilmes Buried Markov Model Pairwise 0.6 0.6 1.3 Structuring Model for Speech Recognition using Buried Markov Model Takayuki Yamamoto, 1 Tetsuya Takiguchi
ΠΕΡΙΕΧΟΜΕΝΑ. Κεφάλαιο 1: Κεφάλαιο 2: Κεφάλαιο 3:
 4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
4 Πρόλογος Η παρούσα διπλωµατική εργασία µε τίτλο «ιερεύνηση χωρικής κατανοµής µετεωρολογικών µεταβλητών. Εφαρµογή στον ελληνικό χώρο», ανατέθηκε από το ιεπιστηµονικό ιατµηµατικό Πρόγραµµα Μεταπτυχιακών
ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ
 ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Τομέας Περιβαλλοντικής Υδραυλικής και Γεωπεριβαλλοντικής Μηχανικής (III) Εργαστήριο Γεωπεριβαλλοντικής Μηχανικής TECHNICAL UNIVERSITY OF CRETE SCHOOL of
ΠΟΛΥΤΕΧΝΕΙΟ ΚΡΗΤΗΣ ΣΧΟΛΗ ΜΗΧΑΝΙΚΩΝ ΠΕΡΙΒΑΛΛΟΝΤΟΣ Τομέας Περιβαλλοντικής Υδραυλικής και Γεωπεριβαλλοντικής Μηχανικής (III) Εργαστήριο Γεωπεριβαλλοντικής Μηχανικής TECHNICAL UNIVERSITY OF CRETE SCHOOL of
Other Test Constructions: Likelihood Ratio & Bayes Tests
 Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
Other Test Constructions: Likelihood Ratio & Bayes Tests Side-Note: So far we have seen a few approaches for creating tests such as Neyman-Pearson Lemma ( most powerful tests of H 0 : θ = θ 0 vs H 1 :
H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά
 ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ ΝΟΣΗΛΕΥΤΙΚΗ 2008, 47(3):367 373 H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά Ανθή Χρυσοστόμου Νοσηλεύτρια ΠΕ, ΜSc, Kέντρο Ελέγχου και Πρόληψης
ΕΡΕΥΝΗΤΙΚΗ ΕΡΓΑΣΙΑ ΝΟΣΗΛΕΥΤΙΚΗ 2008, 47(3):367 373 H επίδραση της γονικής παρουσίας και του παιχνιδιού σε επώδυνες διαδικασίες στα παιδιά Ανθή Χρυσοστόμου Νοσηλεύτρια ΠΕ, ΜSc, Kέντρο Ελέγχου και Πρόληψης
Μηχανισμοί πρόβλεψης προσήμων σε προσημασμένα μοντέλα κοινωνικών δικτύων ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ
 ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΕΠΙΚΟΙΝΩΝΙΩΝ, ΗΛΕΚΤΡΟΝΙΚΗΣ ΚΑΙ ΣΥΣΤΗΜΑΤΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ Μηχανισμοί πρόβλεψης προσήμων σε προσημασμένα μοντέλα κοινωνικών
ΕΘΝΙΚΟ ΜΕΤΣΟΒΙΟ ΠΟΛΥΤΕΧΝΕΙΟ ΣΧΟΛΗ ΗΛΕΚΤΡΟΛΟΓΩΝ ΜΗΧΑΝΙΚΩΝ ΚΑΙ ΜΗΧΑΝΙΚΩΝ ΥΠΟΛΟΓΙΣΤΩΝ ΤΟΜΕΑΣ ΕΠΙΚΟΙΝΩΝΙΩΝ, ΗΛΕΚΤΡΟΝΙΚΗΣ ΚΑΙ ΣΥΣΤΗΜΑΤΩΝ ΠΛΗΡΟΦΟΡΙΚΗΣ Μηχανισμοί πρόβλεψης προσήμων σε προσημασμένα μοντέλα κοινωνικών
Supplementary Information for
 Supplentary Information for Aggregation induced blue-shifted ission molecular picture from QM/MM study Qunyan Wu, a Tian Zhang, a Qian Peng,* b Dong Wang, a and Zhigang Shuai* a a Key Loratory of Organic
Supplentary Information for Aggregation induced blue-shifted ission molecular picture from QM/MM study Qunyan Wu, a Tian Zhang, a Qian Peng,* b Dong Wang, a and Zhigang Shuai* a a Key Loratory of Organic
