SUPPLEMENTARY INFORMATION
|
|
|
- Καλλιόπη Παπαδόπουλος
- 7 χρόνια πριν
- Προβολές:
Transcript
1 SUPPLEMENTARY INFORMATION doi: /nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros Reviriego, Caroline Phipps, Caroline Sinclair, Catherine Ingle, Catherine McCarthy, Catherine Tudor, Cecilia Icoresi Mazzeo, Christopher Isherwood, Christopher Lelliott, Daniel Barrett, David Tino Lafont, Debarati Sethi, Ellen Brown, Emma Siragher, Evelina Miklejewska, Evelyn Grau, Fiona Kussy, Helen Kundi, Jacqueline White, Joanna Bottomley, Jonathan Burvill, Lauren Anthony, Mark Sanderson, Michael Woods, Monika Dabrowska, Radka Platte, Ramiro Ramirez-Solis, Selina Pearson, Simon Maguire, Susana Caetano, Tanya Bayzetinova, Valerie Vancollie & Yvette Hooks Wellcome Trust Sanger Institute, Wellcome Genome Campus, Cambridge CB10 1SA, UK. 1
2 SI Table 1 Results of the experimental metastasis assay from stage 1 of the screen. A mutant (Mut) mouse line progressed to stage 2 if the metastatic ratio was 0.6 or 1.6 and P (Mann- Whitney test), and additional cohorts were tested. Where multiple cohorts were tested: (i) data is shown for the results from the first cohort, (ii) those with a significant results by Integrative Data Analysis (P < 0.01) are in red/green font (depending if decreased/increased metastasis), and (iii) those without significant results by Integrative Data Analysis (P > 0.01) are in blue font. P- and U-value listed in the table is from a Mann-Whitney test. F, female; M, male; Mut, mutant; WT, wildtype. Mutated Gene <Allele> Background Genotype Sex (mean) (SD) (n) (mean) (SD) (n) Mut/WT P-value U-value F02Rik <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F G10Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M C02Rik <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F K13Rik <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M O03Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F A15Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M P04Rik <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M H14Rik <tm2b(komp)wtsi> C57BL/6NTac Homozygote M J05Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M B14Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F K01Rik <tm2a(komp)wtsi> C57BL/6NTac Heterozygote F E06Rik <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote F O20Rik <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M C02Rik <tm1b(komp)wtsi> C57BL/6NTac Homozygote F E04Rik <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F G23Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F B18Rik <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F N02Rik <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M M09Rik <em1wtsi> C57BL/6NTac Homozygote M H13Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M E14Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F A11Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F C13Rik <tm1b(komp)wtsi> C57BL/6NTac Homozygote M A13Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F K21Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F H24Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M E01Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M J08Rik <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M L15Rik <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M P20Rik <em1wtsi> C57BL/6NTac Homozygote M N03Rik <tm2a(komp)wtsi> C57BL/6NTac Homozygote M E20Rik <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M G04Rik <em1wtsi> C57BL/6NTac Homozygote M N21Rik <tm1b(komp)wtsi> C57BL/6NTac Homozygote M C18Rik <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M E15Rik <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M L06Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M A430005L14Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F A430078G23Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote M A730017C20Rik <tm1b(komp)wtsi> C57BL/6NTac Homozygote M A830019P07Rik <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Abcb6 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Abhd14a <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Abhd16a <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Abhd17a <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Abtb2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Acaa1a <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Acbd5 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Acer1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Acot5 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F
3 Actl10 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Actn4 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Actr6 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Adal <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Adamts19 <tm4a(eucomm)wtsi> C57BL/6NTac Homozygote M Adamts3 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Adap1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Adcy2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Adcy9 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Adpgk <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote F Aff3 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Agap1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Ago3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Ahcyl1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Aicda <tm1b(eucomm)hgmu> C57BL/6NTac Homozygote M Akap9 <tm1a(komp)wtsi> C57BL/6Brd-Tyr, C57BL/6Dnk, C57BL/6NTac Heterozygote F Akr1c20 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Akt2 <tm1e(komp)wtsi> 129S5 Homozygote F Aldh1b1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Aldh3b1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Alox12e <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Alpk1 <em2wtsi> C57BL/6NTac Homozygote M Amz2 <tm1e(komp)wtsi> C57BL/6NTac Homozygote M Anapc4 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Ankrd13d <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Ankrd6 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Ankrd9 <tm1(komp)wtsi> C57BL/6NTac Homozygote F Anks1b <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Anks6 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Ano10 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Anxa6 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Anxa9 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Ap2a2 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Ap4e1 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Ap4m1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Apip <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Apol7a <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Apol9b <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Apoo <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Apool <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Arap2 <tm2b(eucomm)wtsi> C57BL/6NTac Homozygote M Arhgap17 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Arhgap22 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Arhgef1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Arhgef38 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Arhgef7 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Arid1b <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Armc7 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Arpc1b <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Arpc3 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Arrdc5 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Arsg <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Art4 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Asb6 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Aspm <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Atad2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Atad3a <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M
4 Atg16l1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Atg16l2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Atp11a <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Atp5e <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Atp5f1 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Atp8b2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Atxn10 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Atxn3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F B9d2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Bach2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Bai1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote F Bap1 <tm1a(eucomm)hgmu> C57BL/6NTac Heterozygote M BC <tm1a(komp)wtsi> C57BL/6NTac Homozygote F BC <tm1a(komp)wtsi> C57BL/6NTac Homozygote F BC <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Bhlhe40 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Bivm <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Bnip2 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Bpgm <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Bpifb1 <tm1e(komp)wtsi> C57BL/6NTac Homozygote M Bpifb5 <tm2a(komp)wtsi> C57BL/6NTac Homozygote F Brcc3 <em1wtsi> C57BL/6NTac Homozygote M Brd2 <em1wtsi> C57BL/6NTac Heterozygote M Brd8 <tm1a(eucomm)wtsi> C57BL/6Brd-Tyr, C57BL/6Dnk, C57BL/6NTac Heterozygote F Cabp1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cadm1 <tm1.2brd> C57BL/6Brd-Tyr, C57BL/6J, 129S5/SvEvBrd Homozygote F Calcoco2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Camkmt <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Camsap3 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Capn11 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Capza2 <tm1(komp)wtsi> C57BL/6NTac Heterozygote M Card9 <tm1a(eucomm)hmgu> C57BL/6NTac Homozygote M Casc4 <tm2b(eucomm)wtsi> C57BL/6NTac Homozygote M Casp12 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Catip <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Cbr2 <tm1a(eucomm)hgmu> C57BL/6NTac Homozygote F Cbx6 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Ccdc122 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Ccdc127 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Ccdc134 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Ccdc159 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Ccdc160 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Ccdc18 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Ccdc24 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Ccdc6 <em1wtsi> C57BL/6NTac Heterozygote M Ccdc69 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Ccl22 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Cd300lg <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Cdca8 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Cdh19 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cdh23 <v> Mixed Homozygote M Cdk4 <R24C/R24/C> C57BL/6 Homozygote M Cdk5rap2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Cdkal1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Cdkn2aipnl <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Cdyl <em1wtsi> C57BL/6NTac Heterozygote M
5 Celf4 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Cenpl <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Cep250 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Cfap61 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Cgrrf1 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Chd9 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Chil4 <tm1b(eucomm)hmgu> C57BL/6NTac Homozygote M Chka <tm2a(komp)wtsi> C57BL/6NTac Heterozygote M Chmp6 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Chst11 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Chtop <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Cir1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Clk1 <tm1a(eucomm)wtsi/ics> C57BL/6NTac Homozygote F Clpp <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cmbl <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Cmip <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Cmtm4 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cmtm5 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Cmtm6 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cnbd1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Cnih3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Cnot4 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Cnot6 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Cog6 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Col24a1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Col4a3bp <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Commd10 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Copg <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Coq4 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Coro1c <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Coro6 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote F Cpsf3 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Cpsf3l <tm2b(eucomm)wtsi> C57BL/6NTac Heterozygote M Cpt2 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Crb2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Creb3l1 <tm1e(eucomm)wtsi> C57BL/6NTac Heterozygote F Crim1 <em1wtsi> C57BL/6NTac Heterozygote M Crip2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Crlf3 <tm1a(komp)wtsi> C57BL/6Brd-Tyr, C57BL/6Dnk, C57BL/6NTac Homozygote F Crls1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Csmd1 <em2wtsi> C57BL/6NTac Homozygote M Csnk1g3 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Ctr9 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Cttnbp2 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Cutal <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Cxcr1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Cxcr2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cyba <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Cybb <tm1din> C57BL/6J Homozygote F Cyfip1 <tm2a(eucomm)wtsi> C57BL/6NTac Heterozygote F Cyfip2 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Cyp11a1 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Cyp20a1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Cyp2b13 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Cyp2r1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Cyp3a11 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F D630023F18Rik <tm1b(komp)wtsi> C57BL/6NTac Homozygote M
6 D7Ertd443e <tm2a(komp)wtsi> C57BL/6NTac Homozygote M D930028M14Rik <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Daam2 <tm1a(komp)wts> C57BL/6NTac Homozygote F Dact3 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Daf2 <tm1a(komp)wts> C57BL/6NTac Homozygote F Dap <tm1a(komp)mbp> C57BL/6NTac Homozygote M Dapk2 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Dbn1 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Dcaf11 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dcbld2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dcdc2b <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Dcdc2c <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dclk1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Dcn <em2wtsi> C57BL/6NTac Homozygote M Dctn1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Dcun1d3 <em2wtsi> C57BL/6NTac Heterozygote M Dcx <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Ddah1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Ddhd1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Ddx42 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Ddx51 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Ddx55 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Def6 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Defb14 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Defb22 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dennd1b <tm1a(eucomm)hmgu> C57BL/6NTac Homozygote F Dennd1c <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Dennd4c <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Denr <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Dffb <em1wtsi> C57BL/6NTac Homozygote M Dhodh <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Dhps <tm2a(eucomm)wtsi> C57BL/6NTac Heterozygote M Dhrs2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dhx33 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Dhx35 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Dip2a <tm2b(komp)wtsi> C57BL/6NTac Homozygote F Dlg2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Dlg3 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Dlg4 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote M Dmgdh <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dmxl2 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Dnah17 <tm1e(komp)wtsi> C57BL/6NTac Homozygote M Dnajc8 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Dnase1l2 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Dnmt3a <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dnpep <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote M Donson <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Dopey2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Dph2 <tm2(eucomm)wtsi> C57BL/6NTac Heterozygote M Dph6 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dpm1 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Dppa1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dpy30 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Dsc2 <tm1e(komp)wtsi> C57BL/6NTac Homozygote F Dscc1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Dsg1b <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Dusp5 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F
7 Dynlrb1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Dynlrb2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Eci3 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Edc4 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Efna1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Ehbp1l1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Eif2b2 <tm2a(komp)wtsi> C57BL/6NTac Heterozygote F Eif3h <tm1a(eucomm)hmgu> C57BL/6NTac Heterozygote M Elac2 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Ell2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Elmo1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Enc1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Endou <tm1> C57BL/6NTac Homozygote F Enthd2 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Entpd1 <tm1a(eucomm)wtsi/hmgu> C57BL/6Dnk Homozygote F Entpd6 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Erlin2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Erp44 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Esco1 <tm1(komp)wtsi> C57BL/6NTac Homozygote M Espn <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Evi5 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Exoc3l2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Exosc9 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Expi <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Fads3 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Fam122c <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Fam160a1 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Fam163a <tm2b(komp)wtsi> C57BL/6NTac Homozygote M Fam175b <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Fam21 <tm2b(komp)wtsi> C57BL/6NTac Heterozygote M Fam212b <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Fam46c <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Fam47e <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Fam69a <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Fam92a <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Fanci <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Fbf1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Fbxo33 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Fbxo7 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Fbxw26 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Fdft1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Fggy <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Fkbp3 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Fnip2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Frmd7 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Frrs1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Fryl <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Fut2 <em1wtsi> C57BL/6NTac Homozygote M Fut8 <em1wtsi> C57BL/6NTac Heterozygote F Fxyd3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Fzd6 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M G3bp2 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Galnt18 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Galntl5 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F
8 Gas2l2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Gbf1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Gbp5 <em1wtsi> C57BL/6NTac Homozygote M Gda <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Gdpd2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Gimap6 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Gldc <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Glg1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Glo1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Glt8d2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Glyatl3 <em1wtsi> C57BL/6NTac Homozygote M Gm12253 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Gm13119 <tm2b(komp)wtsi> C57BL/6NTac Homozygote M Gm13125 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Gm16379 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Gm16432 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Gm5544 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Gm648 <tm1e(komp)wtsi> C57BL/6NTac Homozygote F Gm6578 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Gmds <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Gmnc <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Gnb3 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Gpatch1 <em2wtsi> C57BL/6NTac Heterozygote M Gpbp1l1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Gpc5 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Gpr107 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Gpr133 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Gpr152 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Gpr35 <tm1b(eucomm)hmgu> C57BL/6NTac Homozygote M Grb7 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Grm8 <tm2a(komp)wtsi> C57BL/6NTac Homozygote F Grsf1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Gsto2 <tm2a(komp)wtsi> C57BL/6NTac Homozygote F Gtf2a1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Gtf2h2 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Gtpbp3 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M H2-Eb1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Hacl1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Hao2 <em1wtsi> C57BL/6NTac Homozygote M Hbs1l <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Hdac8 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Hecw2 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Herc1 <em4wtsi> (T1696G point mutation) C57BL/6NTac Homozygote F Hibadh <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Hmgxb3 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Homez <tm1e(komp)wtsi> C57BL/6NTac Homozygote M Hrsp12 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Hsp90aa1 <tm1(komp)wtsi> C57BL/6NTac Homozygote F Ido1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Ifi27 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Ifitm3 <tm1masu> C57/129 mixed Homozygote F Ifitm6 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Ifnar1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote F Ifnlr1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Ift140 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F
9 Ift80 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Ighm <tm1(cgn)> C57BL/6J Homozygote F Il10rb <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Il17re <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Il18rap <em1wtsi> C57BL/6NTac Homozygote M Il1r2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Il22ra1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Il23r <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Ildr1 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Ints2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Irak1 <tm1a(eucomm)hgmu> C57BL/6NTac Homozygote M Irf1 <tm1a(eucomm)wtsi> C57BL/6Brd-Tyr, C57BL/6Dnk, C57BL/6NTac Homozygote F Irf5 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote F Irf7 <tm1(komp)wtsi> C57BL/6NTac Homozygote F Isg20 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Itln1 <em1wtsi> C57BL/6NTac Homozygote M Izumo1r <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Jhdm1d <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Jmjd1c <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Josd1 <em2wtsi> C57BL/6NTac Homozygote F Kcnh4 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Kcnip4 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Kdm4d <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote F Kera <em1wtsi> C57BL/6NTac Homozygote M Kif13b <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Kif18b <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Kif24 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Kif3b <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Kifap3 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Klc2 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote M Klf17 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Klhl18 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Klhl21 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Klhl30 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Klk5 <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Klk9 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Klrb1a <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Kptn <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Krt31 <tm1e(komp)wtsi> C57BL/6NTac Homozygote M Krt7 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F L3mbtl2 <tm1e(eucomm)wtsi> C57BL/6NTac Heterozygote M Lacc1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Lars <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Lce1m <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Lce3c <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote F Ldhb <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Ldlrad4 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Leo1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Leprot <tm2a(komp)wtsi> C57BL/6NTac Homozygote F Lgals7 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Lix1l <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Lmna <G609G> C57BL/6NTac Homozygote F Lonrf3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Lpxn <tm1b(eucomm)hgmu> C57BL/6NTac Homozygote M Lrig1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Lrrc23 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Lrrc71 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M
10 Lrrc8d <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Lyplal1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Lyrm9 <tm1a(komp)mbp> C57BL/6NTac Homozygote M Macrod1 <em1wtsi> C57BL/6NTac Homozygote M Mamstr <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Man2b2 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Map2k7 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Map3k1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Mast2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Mau2 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Mbl2 <em1wtsi> C57BL/6NTac Homozygote M Mcf2l <tm1a(eucomm)hmgu> C57BL/6NTac Homozygote F Med22 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Med23 <em1wtsi> C57BL/6NTac Heterozygote M Medag <tm2b(komp)wtsi> C57BL/6NTac Homozygote F Mep1a <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Metrnl <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Mettl24 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Mgat4c <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Micu2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Mier1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F mir122 <tm1brd> C57BL/6NTac Homozygote M mir145 <tm1brd> C57BL/6NTac Heterozygote F mir210 <tm1brd> C57BL/6NTac Homozygote F mir211 <tm1brd> C57BL/6NTac Homozygote M mir32 <tm1brd> C57BL/6NTac Homozygote M mir342 <tm1brd> C57BL/6NTac Homozygote M mir34a <tm1brd> C57BL/6NTac Homozygote F mir96 <tm2.1wtsi> C57BL/6NTac Heterozygote M mir-96, mir-183 ('cluster6n1') <tm1brd> C57BL/6NTac Homozygote M mir-25, mir-93, mir-106b ('cluster5n1') <tm1brd> C57BL/6NTac Homozygote M mir-18b, mir-106a ('clusterxn1') <tm1brd> C57BL/6NTac Homozygote M Mkrn2 <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Mospd1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Mpg <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Mrm1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote F Mroh4 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Mroh9 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Mrps21 <tm1e(eucomm)wtsi> C57BL/6NTac Heterozygote M Mrps5 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Mtmr1 <tm2a(eucomm)wtsi> C57BL/6NTac Homozygote M Mtss1l <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Mybphl <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Myd88 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Myh14 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Myh7b <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Myo10 <tm2(komp)wtsi> C57BL/6NTac Homozygote M Myo7a <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Myo9a <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Mysm1 <tm1a(komp)wtsi> C57BL/6Dnk, C57BL/6NTac Heterozygote F Myt1l <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M N6amt2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Naaa <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Nacad <tm1(komp)wtsi> C57BL/6NTac Homozygote F Nacc2 <tm2b(eucomm)wtsi> C57BL/6NTac Homozygote M Nadk2 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Nat10 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F
11 Natd1 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Nbeal2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Ncf2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Ndufb8 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Nebl <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Nek3 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Nek9 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Nelfe <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Nfkb1 <tm1a(komp)wtsi> C57BL/6Brd-Tyr, C57BL/6Dnk, C57BL/6NTac Homozygote F Nfkbil1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Nfu1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Ngrn <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Nhlrc2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Nhp2 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Ninj2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Nipsnap3a <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Nme4 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Nmrk1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Npat <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Nrbp1 <tm3a(eucomm)wtsi> C57BL/6NTac Heterozygote F Nrde2 <tm2a(komp)wtsi> C57BL/6NTac Heterozygote F Nubpl <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M cc18 Nudt12 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Nup85 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Nutm2 <tm2b(eucomm)wtsi> C57BL/6NTac Homozygote F Nxn <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Oaf <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Oas1g <tm3e(komp)wtsi> C57BL/6NTac Homozygote F Ocm <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote F Ogdh <tm1e(komp)wtsi> C57BL/6NTac Heterozygote M Olfr168 <KO> C57BL/6NTac Heterozygote M Oog2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Orc1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Os9 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Osbpl3 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Otub2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Otud7b <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Oxr1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Pabpc4 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Pabpn1l <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Pam16 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M BC <em1(gb)paxx> C57BL/6NTac Homozygote M Pced1a <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote F Pdcd2 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Pdia4 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Pdk3 <tm2a(komp)wtsi> C57BL/6NTac Homozygote F Pds5b <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Pdxk <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Pdzd3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Pdzk1 <tm2b(eucomm)wtsi> C57BL/6NTac Homozygote M Pear1 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Pepd <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Pfkfb4 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Pgap2 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Pigf <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Pigl <tm1b(komp)wtsi> C57BL/6NTac Heterozygote F Pik3cg <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M
12 Pitrm1 <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Pkhd1 <del2> C57BL/6, 129S6/SvEvTac Homozygote F Pla2g6 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Pld3 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote M Plekhg1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Plet1 <tm2b(komp)wtsi> C57BL/6NTac Homozygote F Pls3 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Plscr2 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Plxna2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Plxnb3 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Polb <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Pold3 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Polr3f <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Polr3g <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Pop4 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Ppil3 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Ppp6c <tm1a(eucomm)hgmu> C57BL/6NTac Heterozygote F Praf2 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote F Prame <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Prkab1 <tm1b(komp)wtsi> C57BL/6NTac Homozygote F Prrc2b <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Prrg2 <tm1b(eucomm)wtsi> C57BL/6NTac Homozygote M Prrt2 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Prss52 <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Prss53 <em2wtsi> C57BL/6NTac Homozygote M Psat1 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Dlg4 <tm1grnt> C57BL/6;129 Homozygote M Psph <tm1a(eucomm)hmgu> C57BL/6NTac Heterozygote M Pth <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Pth1r <tm1a(eucomm)hmgu> C57BL/6NTac Heterozygote M Pth2r <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Pthlh <tm1a(komp)wtsi> C57BL/6NTac Heterozygote M Ptprd <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Pus10 <tm1a(eucomm)hgmu> C57BL/6NTac Homozygote M Pwp1 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Pycr2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Rab15 <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Rab17 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Rab21 <tm1b(komp)wtsi> C57BL/6NTac Heterozygote M Rab5c <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Rala <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote M Ralgapb <tm1a(komp)wtsi> C57BL/6NTac Heterozygote F Ralgps2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Raph1 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote F Rasal2 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Rasgrp4 <tm2a(komp)wtsi> C57BL/6NTac Homozygote M Rassf1a <tm1.2brd> C57BL/6Brd-Tyr, C57BL/6J, 129S5/SvEvBrd Homozygote F Rbak <tm1b(komp)wtsi> C57BL/6NTac Homozygote M Rbm14 <tm1a(komp)wtsi> C57BL/6NTac Homozygote M Rbm33 <tm1b(eucomm)wtsi> C57BL/6NTac Heterozygote M Rbm47 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote M Rbmx <tm2a(komp)wtsi> C57BL/6NTac Homozygote F Rcor2 <tm1a(eucomm)wtsi> C57BL/6NTac Heterozygote F Reg3d <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Reg3g <tm1a(komp)wtsi> C57BL/6NTac Homozygote F Repin1 <tm1a(eucomm)wtsi> C57BL/6NTac Homozygote F Rftn2 <tm1e(eucomm)wtsi> C57BL/6NTac Homozygote F
IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion
 Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
Supplementary figures
 A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
A Supplementary figures a) DMT.BG2 0.87 0.87 0.72 20 40 60 80 100 DMT.EG2 0.93 0.85 20 40 60 80 EMT.MG3 0.85 0 20 40 60 80 20 40 60 80 100 20 40 60 80 100 20 40 60 80 EMT.G6 DMT/EMT b) EG2 0.92 0.85 5
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Nature Medicine doi: /nm.2772
 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Online Content
 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
NCBI ID Symbol 2 A2M A4GALT A4GNT 8086 AAAS 13 AADAC AADACL AAGAB 15 AANAT 16 AARS AARS AASS 18 ABAT 19 ABCA1
 NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
< (0.999) Graft (0.698) (0.483) <0.001 (0.698) (<0.001) (<0.001) 3 months (0.999) (0.483) (<0.001) 6 months (<0.
 Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Supplementary table 1. Correlation of endothelial cell density among graft, 3, 6, and 12 months after Descemet s automated stripping endothelial keratoplasty. Graft 3 months 6 months 12 months Graft
Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal
 Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplementary Materials for
 www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
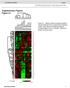 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
Molecular evolutionary dynamics of respiratory syncytial virus group A in
 Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya James R. Otieno 1#, Charles N. Agoti 1, 2, Caroline W. Gitahi 1, Ann Bett 1, Mwanajuma Ngama
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis
 S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
500 New CUSABIO Antibodies at MedStore!
 BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
 Νόσος Parkinson-Νέοι ορίζοντες Ξηροµερήσιου Γεωργία Νευρολόγος-Μέλος ερευνητικής οµάδας Νευρογενετικής Αίτια της νόσου Parkinson Περιβαλλοντικοί παράγοντες Γενετικοί παράγοντες Γενετική βλάβη (παθογόνα
Νόσος Parkinson-Νέοι ορίζοντες Ξηροµερήσιου Γεωργία Νευρολόγος-Μέλος ερευνητικής οµάδας Νευρογενετικής Αίτια της νόσου Parkinson Περιβαλλοντικοί παράγοντες Γενετικοί παράγοντες Γενετική βλάβη (παθογόνα
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells
 Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
A strategy for the identification of combinatorial bioactive compounds. contributing to the holistic effect of herbal medicines
 1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
1 2 Supplementary information 3 4 A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines 5 6 Fang Long 1, Hua Yang 1, Yanmin Xu,
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma. Supplement Figure 1. Regulation of PGC1α expression in cultured podocytes.
 Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
Supplementary Figure 1.
 Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)]
![Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)] Technical Information T-9100 SI. Suva. refrigerants. Thermodynamic Properties of. Suva Refrigerant [R-410A (50/50)]](/thumbs/89/98928029.jpg) d Suva refrigerants Technical Information T-9100SI Thermodynamic Properties of Suva 9100 Refrigerant [R-410A (50/50)] Thermodynamic Properties of Suva 9100 Refrigerant SI Units New tables of the thermodynamic
d Suva refrigerants Technical Information T-9100SI Thermodynamic Properties of Suva 9100 Refrigerant [R-410A (50/50)] Thermodynamic Properties of Suva 9100 Refrigerant SI Units New tables of the thermodynamic
Supporting Information
 Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1δ/ε Inhibitor Germana Esposito, Marie-Lise Bourguet-Kondracki, * Linh H. Mai, Arlette Longeon, Roberta Teta, Laurent Meijer, Rob Van Soest,
Aquinas College. Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET
 Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Aquinas College Edexcel Mathematical formulae and statistics tables DO NOT WRITE ON THIS BOOKLET Pearson Edexcel Level 3 Advanced Subsidiary and Advanced GCE in Mathematics and Further Mathematics Mathematical
Analysis Summary: Any Allergy
 Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue.
 Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
Supplementary Table 1. Primers used for RT-qPCR analysis of striatal and nigral tissue. Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) Dopaminergic Markers TH CTG GCC ATT GAT GTA CTG GA ACA CAC ATG GGA
DuPont Suva 95 Refrigerant
 Technical Information T-95 SI DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 SI DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
DuPont Suva 95 Refrigerant
 Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-95 ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 95 Refrigerant (R-508B) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Math 6 SL Probability Distributions Practice Test Mark Scheme
 Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
Math 6 SL Probability Distributions Practice Test Mark Scheme. (a) Note: Award A for vertical line to right of mean, A for shading to right of their vertical line. AA N (b) evidence of recognizing symmetry
CD4 + T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2
 Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland
Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
UMI Number: All rights reserved
 UMI Number: 3408360 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send
UMI Number: 3408360 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send
DuPont Suva. DuPont. Thermodynamic Properties of. Refrigerant (R-410A) Technical Information. refrigerants T-410A ENG
 Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
Technical Information T-410A ENG DuPont Suva refrigerants Thermodynamic Properties of DuPont Suva 410A Refrigerant (R-410A) The DuPont Oval Logo, The miracles of science, and Suva, are trademarks or registered
; +302 ; +313; +320,.
 1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
1.,,*+, - +./ +/2 +, -. ; +, - +* cm : Key words: snow-water content, surface soil, snow type, water permeability, water retention +,**. +,,**/.. +30- +302 ; +302 ; +313; +320,. + *+, *2// + -.*, **. **+.,
Supplementary Figure 1
 NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
NSC N2d N5d TNFRSFa TNFRSFb RIPK RelA Madd I-TRAF/TANK GAPDH γ-actin Supplementary Figure A 2 hrs Day : Cell seeding Day 9: +ng/ml TNF-α Day 9: Fixation B Merge Nestin PH-3 Hoechst Acute TNF-α Control
 Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
FORMULAS FOR STATISTICS 1
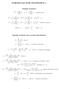 FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
FORMULAS FOR STATISTICS 1 X = 1 n Sample statistics X i or x = 1 n x i (sample mean) S 2 = 1 n 1 s 2 = 1 n 1 (X i X) 2 = 1 n 1 (x i x) 2 = 1 n 1 Xi 2 n n 1 X 2 x 2 i n n 1 x 2 or (sample variance) E(X)
Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks. Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks
 Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Group 1 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Group 2 Methotrexate 7.5 mg/week, increased to 15 mg/week after 4 weeks Sulfasalazine 2000-3000 mg/day Leflunomide 20 mg/day Infliximab
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Single-site association results for 136 SCARB1 genotyped variants with HDL-C.
 Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
Table S9. Single-site association results for 136 SCARB1 genotyped variants with HDL-C. SNP Name a SNP ID b Chr12 Position c Location Amino Acid Change RegDB Score d MA, MAF Genotype Genotype Count Adjusted
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 19/5/2007
 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Αν κάπου κάνετε κάποιες υποθέσεις να αναφερθούν στη σχετική ερώτηση. Όλα τα αρχεία που αναφέρονται στα προβλήματα βρίσκονται στον ίδιο φάκελο με το εκτελέσιμο
Supporting Information
 Supporting Information Wiley-VCH 27 69451 Weinheim, Germany Supplementary Figure 1. Synthetic results as detected by XRD (Cu-Kα). Simulation pattern of MCM-68 Relative Intensity / a.u. YNU-2P Conventional
Supporting Information Wiley-VCH 27 69451 Weinheim, Germany Supplementary Figure 1. Synthetic results as detected by XRD (Cu-Kα). Simulation pattern of MCM-68 Relative Intensity / a.u. YNU-2P Conventional
ΚΥΠΡΙΑΚΗ ΕΤΑΙΡΕΙΑ ΠΛΗΡΟΦΟΡΙΚΗΣ CYPRUS COMPUTER SOCIETY ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 11/3/2006
 ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 11/3/26 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα μικρότεροι το 1 εκτός αν ορίζεται διαφορετικά στη διατύπωση
ΠΑΓΚΥΠΡΙΟΣ ΜΑΘΗΤΙΚΟΣ ΔΙΑΓΩΝΙΣΜΟΣ ΠΛΗΡΟΦΟΡΙΚΗΣ 11/3/26 Οδηγίες: Να απαντηθούν όλες οι ερωτήσεις. Ολοι οι αριθμοί που αναφέρονται σε όλα τα ερωτήματα μικρότεροι το 1 εκτός αν ορίζεται διαφορετικά στη διατύπωση
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
Daewoo Technopark A-403, Dodang-dong, Wonmi-gu, Bucheon-city, Gyeonggido, Korea LM-80 Test Report
 LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
LM-80 Test Report Approved Method: Measuring Lumen Maintenance of LED Light Sources Project Number: KILT1212-U00216 Date: September 17 th, 2013 Requested by: Dongbu LED Co., Ltd 90-1, Bongmyeong-Ri, Namsa-Myeon,
The effect of curcumin on the stability of Aβ. dimers
 The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
The effect of curcumin on the stability of Aβ dimers Li Na Zhao, See-Wing Chiu, Jérôme Benoit, Lock Yue Chew,, and Yuguang Mu, School of Physical and Mathematical Sciences, Nanyang Technological University,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
5- CACGAAACTACCTTCAACTCC-3 beta actin-r 5- CATACTCCTGCTTGCTGATC-3 GAPDH-F GAPDH-R
 Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Matrices and Determinants
 Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Matrices and Determinants SUBJECTIVE PROBLEMS: Q 1. For what value of k do the following system of equations possess a non-trivial (i.e., not all zero) solution over the set of rationals Q? x + ky + 3z
Burden of Disease Variants in Participants of the Long Life Family Study. Supplemental Online Material
 Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Cable Systems - Postive/Negative Seq Impedance
 Cable Systems - Postive/Negative Seq Impedance Nomenclature: GMD GMR - geometrical mead distance between conductors; depends on construction of the T-line or cable feeder - geometric mean raduius of conductor
Cable Systems - Postive/Negative Seq Impedance Nomenclature: GMD GMR - geometrical mead distance between conductors; depends on construction of the T-line or cable feeder - geometric mean raduius of conductor
± 20% ± 5% ± 10% RENCO ELECTRONICS, INC.
 RL15 RL16, RL17, RL18 MINIINDUCTORS CONFORMALLY COATED MARKING The nominal inductance is marked by a color code as listed in the table below. Color Black Brown Red Orange Yellow Green Blue Purple Grey
RL15 RL16, RL17, RL18 MINIINDUCTORS CONFORMALLY COATED MARKING The nominal inductance is marked by a color code as listed in the table below. Color Black Brown Red Orange Yellow Green Blue Purple Grey
Supplementary Information 1.
 Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
Supplementary Information 1. Fig. S1. Correlations between litter-derived-c and N (percent of initial input) and Al-/Fe- (hydr)oxides dissolved by ammonium oxalate (AO); a) 0 10 cm; b) 10 20 cm; c) 20
ΠΕΡΙΓΡΑΦΙΚΗ και ΕΠΑΓΩΓΙΚΗ ΣΤΑΤΙΣΤΙΚΗ
 ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΠΕΡΙΓΡΑΦΙΚΗ και ΕΠΑΓΩΓΙΚΗ ΣΤΑΤΙΣΤΙΚΗ Εισήγηση 5Α: ΠΑΡΑΜΕΤΡΙΚΟ Χ 2 Διδάσκων: Δαφέρμος Βασίλειος ΤΜΗΜΑ ΠΟΛΙΤΙΚΗΣ ΕΠΙΣΤΗΜΗΣ ΣΧΟΛΗΣ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ Άδειες Χρήσης
ΕΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΠΑΝΕΠΙΣΤΗΜΙΟ ΚΡΗΤΗΣ ΠΕΡΙΓΡΑΦΙΚΗ και ΕΠΑΓΩΓΙΚΗ ΣΤΑΤΙΣΤΙΚΗ Εισήγηση 5Α: ΠΑΡΑΜΕΤΡΙΚΟ Χ 2 Διδάσκων: Δαφέρμος Βασίλειος ΤΜΗΜΑ ΠΟΛΙΤΙΚΗΣ ΕΠΙΣΤΗΜΗΣ ΣΧΟΛΗΣ ΚΟΙΝΩΝΙΚΩΝ ΕΠΙΣΤΗΜΩΝ Άδειες Χρήσης
[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,
![[11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13, [11].,, , 316 6, ,., 15.5%, 9.8%, 2006., IDF,, ,500, 2,830.,, ,200.,,, β, [12]. 90% 2,,,,, [13-15].,, [13,](/thumbs/89/98694254.jpg) in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
in vitro αα In Vitro α-glucosidase and α-amylase Inhibitory Effects of Herbal Tea Leaves from Yacon (Smallanthus sonchifolius) 1, 1, 1, 2, 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 2 1, 2, *, 1 2, 3 Yuto Ueda 1, Shintaro
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) CPh 3 as a functional group in P-heterocyclic
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information (ESI) CPh 3 as a functional group in P-heterocyclic
Si + Al Mg Fe + Mn +Ni Ca rim Ca p.f.u
 .6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
.6.5. y = -.4x +.8 R =.9574 y = - x +.14 R =.9788 y = -.4 x +.7 R =.9896 Si + Al Fe + Mn +Ni y =.55 x.36 R =.9988.149.148.147.146.145..88 core rim.144 4 =.6 ±.6 4 =.6 ±.18.84.88 p.f.u..86.76 y = -3.9 x
Supporting Information for: electron ligands: Complex formation, oxidation and
 Supporting Information for: The diverse reactions of PhI(OTf) 2 with common 2- electron ligands: Complex formation, oxidation and oxidative coupling Thomas P. Pell, Shannon A. Couchman, Sara Ibrahim, David
Supporting Information for: The diverse reactions of PhI(OTf) 2 with common 2- electron ligands: Complex formation, oxidation and oxidative coupling Thomas P. Pell, Shannon A. Couchman, Sara Ibrahim, David
Simon et al. Supplemental Data Page 1
 Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Simon et al. Supplemental Data Page 1 Supplemental Data Acute hemodynamic effects of inhaled sodium nitrite in pulmonary hypertension associated with heart failure with preserved ejection fraction Short
Iodine-catalyzed synthesis of sulfur-bridged enaminones and chromones via double C(sp 2 )-H thiolation
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2017 Iodine-catalyzed synthesis of sulfur-bridged enaminones and chromones via
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2017 Iodine-catalyzed synthesis of sulfur-bridged enaminones and chromones via
ΔΘΝΙΚΗ ΥΟΛΗ ΓΗΜΟΙΑ ΓΙΟΙΚΗΗ ΙΗ ΔΚΠΑΙΓΔΤΣΙΚΗ ΔΙΡΑ
 Δ ΔΘΝΙΚΗ ΥΟΛΗ ΓΗΜΟΙΑ ΓΙΟΙΚΗΗ ΙΗ ΔΚΠΑΙΓΔΤΣΙΚΗ ΔΙΡΑ ΣΜΗΜΑ ΠΔΡΙΦΔΡΔΙΑΚΗ ΓΙΟΙΚΗΗ ΣΔΛΙΚΗ ΔΡΓΑΙΑ Θέκα: Αμηνιφγεζε κίαο δηαπξαγκάηεπζεο. Μειέηε Πεξίπησζεο: Ζ αλέγεξζε ηεο Νέαο Δζληθήο Λπξηθήο θελήο, ηεο Νέαο
Δ ΔΘΝΙΚΗ ΥΟΛΗ ΓΗΜΟΙΑ ΓΙΟΙΚΗΗ ΙΗ ΔΚΠΑΙΓΔΤΣΙΚΗ ΔΙΡΑ ΣΜΗΜΑ ΠΔΡΙΦΔΡΔΙΑΚΗ ΓΙΟΙΚΗΗ ΣΔΛΙΚΗ ΔΡΓΑΙΑ Θέκα: Αμηνιφγεζε κίαο δηαπξαγκάηεπζεο. Μειέηε Πεξίπησζεο: Ζ αλέγεξζε ηεο Νέαο Δζληθήο Λπξηθήο θελήο, ηεο Νέαο
An experimental and theoretical study of the gas phase kinetics of atomic chlorine reactions with CH 3 NH 2, (CH 3 ) 2 NH, and (CH 3 ) 3 N
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 An experimental and theoretical study of the gas phase kinetics of atomic chlorine
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 An experimental and theoretical study of the gas phase kinetics of atomic chlorine
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models
 Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Supplementary Material for The Cusp Catastrophe Model as Cross-Sectional and Longitudinal Mixture Structural Equation Models Sy-Miin Chow Pennsylvania State University Katie Witkiewitz University of New
Strain gauge and rosettes
 Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
Strain gauge and rosettes Introduction A strain gauge is a device which is used to measure strain (deformation) on an object subjected to forces. Strain can be measured using various types of devices classified
SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in
 1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
Reminders: linear functions
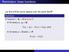 Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Reminders: linear functions Let U and V be vector spaces over the same field F. Definition A function f : U V is linear if for every u 1, u 2 U, f (u 1 + u 2 ) = f (u 1 ) + f (u 2 ), and for every u U
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
SUPPLEMENTARY INFORMATION
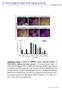 doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ
 ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ Μαυρόγιαννη Γεωργία 1, Καραντζούλα Ευαγγελία 2, Τουλιά Γεωργία 3, Κυριακόπουλος Βασίλης 4, Καδδά Όλγα 5 EΡΕΥΝΑ
ΚΕΡΚΙΔΙΚΗ ΚΑΙ ΜΗΡΙΑΙΑ ΠΡΟΣΠΕΛΑΣΗ ΣΕ ΣΤΕΦΑΝΙΟΓΡΑΦΙΕΣ ΚΑΙ ΑΓΓΕΙΟΠΛΑΣΤΙΚΕΣ. ΜΙΑ ΣΥΓΚΡΙΤΙΚΗ ΜΕΛΕΤΗ Μαυρόγιαννη Γεωργία 1, Καραντζούλα Ευαγγελία 2, Τουλιά Γεωργία 3, Κυριακόπουλος Βασίλης 4, Καδδά Όλγα 5 EΡΕΥΝΑ
SMD Wire Wound Ferrite Chip Inductors - LS Series. LS Series. Product Identification. Shape and Dimensions / Recommended Pattern LS0402/0603/0805/1008
 SMD Wire Wound Ferrite Chip Inductors - LS Series LS Series LS Series is the newest in open type ferrite wire wound chip inductors. The wire wound ferrite construction supports higher SRF, lower DCR and
SMD Wire Wound Ferrite Chip Inductors - LS Series LS Series LS Series is the newest in open type ferrite wire wound chip inductors. The wire wound ferrite construction supports higher SRF, lower DCR and
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis
 Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Does anemia contribute to end-organ dysfunction in ICU patients Statistical Analysis Xue Han, MPH and Matt Shotwell, PhD Department of Biostatistics Vanderbilt University School of Medicine March 14, 2014
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
Divergent synthesis of various iminocyclitols from D-ribose
 Electronic Supplementary Material (ESI) for rganic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 205 Divergent synthesis of various iminocyclitols from D-ribose Ramu Petakamsetty,
Electronic Supplementary Material (ESI) for rganic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 205 Divergent synthesis of various iminocyclitols from D-ribose Ramu Petakamsetty,
Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent
 Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Διπλωματική Εργασία. Μελέτη των μηχανικών ιδιοτήτων των stents που χρησιμοποιούνται στην Ιατρική. Αντωνίου Φάνης
 Διπλωματική Εργασία Μελέτη των μηχανικών ιδιοτήτων των stents που χρησιμοποιούνται στην Ιατρική Αντωνίου Φάνης Επιβλέπουσες: Θεοδώρα Παπαδοπούλου, Ομότιμη Καθηγήτρια ΕΜΠ Ζάννη-Βλαστού Ρόζα, Καθηγήτρια
Διπλωματική Εργασία Μελέτη των μηχανικών ιδιοτήτων των stents που χρησιμοποιούνται στην Ιατρική Αντωνίου Φάνης Επιβλέπουσες: Θεοδώρα Παπαδοπούλου, Ομότιμη Καθηγήτρια ΕΜΠ Ζάννη-Βλαστού Ρόζα, Καθηγήτρια
Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein
 1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
Electronic Supplementary Information:
 Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
Electronic Supplementary Information: 2 6 5 7 2 N S 3 9 6 7 5 2 S N 3 9 Scheme S. Atom numbering for thione and thiol tautomers of thioacetamide 3 0.6 0. absorbance 0.2 0.0 0.5 0.0 0.05 0.00 N 2 Ar km/mol
k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +
[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R + k A = [k, k]( )[a 1, a 2 ] = [ka 1,ka 2 ] 4For the division of two intervals of confidence in R +](/thumbs/73/69566903.jpg) Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Chapter 3. Fuzzy Arithmetic 3- Fuzzy arithmetic: ~Addition(+) and subtraction (-): Let A = [a and B = [b, b in R If x [a and y [b, b than x+y [a +b +b Symbolically,we write A(+)B = [a (+)[b, b = [a +b
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Engineering Tunable Single and Dual Optical. Emission from Ru(II)-Polypyridyl Complexes. Through Excited State Design
 Engineering Tunable Single and Dual Optical Emission from Ru(II)-Polypyridyl Complexes Through Excited State Design Supplementary Information Julia Romanova 1, Yousif Sadik 1, M. R. Ranga Prabhath 1,,
Engineering Tunable Single and Dual Optical Emission from Ru(II)-Polypyridyl Complexes Through Excited State Design Supplementary Information Julia Romanova 1, Yousif Sadik 1, M. R. Ranga Prabhath 1,,
