Comparative transcriptome analysis of peripheral blood mononuclear cells in
|
|
|
- Ἡρωδιάς Ζάχος
- 6 χρόνια πριν
- Προβολές:
Transcript
1 Comparative transcriptome analysis of peripheral blood mononuclear cells in hepatitis B-related acute-on-chronic liver failure Qian Zhou, Wenchao Ding, Longyan Jiang, Jiaojiao Xin, Tianzhou Wu, Dongyan Shi, Jing Jiang, Hongcui Cao, Lanjuan Li, Jun Li Contents Supplemental Table S1. Supplemental Table S2. Supplemental Table S4. Supplemental Table S5.
2 Supplemental Table S1. The clinical characteristics of the patients in the sequencing group. Patients Age Sex ALB ALT AST TB Sodium HBV-DNA (years) (F/M) (g/l) (U/L) (U/L) Cr (μmol/l) INR (μmol/l) (μmol/l) log10(iu/ml) MELD Healthy-1 26 F NA 5.64 Healthy-2 28 F NA 6.84 Healthy-3 29 F NA NA NA Healthy-4 42 M NA NA NA CHB-1 51 M CHB-2 41 M CHB-3 39 M CHB-4 32 M ACLF-1 48 M ACLF-2 54 M ACLF-3 28 M ACLF-4 34 M
3 Supplemental Table S2. The clinical characteristics of the patients in the validation group. Patients Age ALB ALT AST TB Cr Sodium HBV-DNA Sex (F/M) (years) (g/l) (U/L) (U/L) INR (μmol/l) (μmol/l) (μmol/l) log10(iu/ml) MELD Healthy-5 34 M NA NA NA NA Healthy-6 30 F NA NA NA Healthy-7 27 F NA NA NA Healthy-8 27 F NA NA NA Healthy-9 31 F NA NA NA Healthy M NA NA NA Healthy F NA NA NA NA NA Healthy F NA NA NA Healthy F NA NA NA Healthy M NA NA NA Healthy M NA NA NA Healthy M NA NA NA Healthy F NA NA NA Healthy F NA NA NA Healthy M NA NA NA Healthy M NA NA NA Healthy M NA NA NA Healthy M NA NA NA NA NA Healthy M NA NA NA NA NA Healthy F NA NA NA CHB-5 65 F CHB-6 69 F CHB-7 48 M
4 CHB-8 41 M CHB-9 40 M CHB F CHB M CHB F CHB F CHB M CHB M CHB M CHB F CHB F CHB M CHB M CHB M CHB F CHB M CHB M HBV-ACLF-5 60 F HBV-ACLF-6 30 M HBV-ACLF-7 41 M NA HBV-ACLF-8 47 M HBV-ACLF-9 47 M HBV-ACLF M HBV-ACLF M HBV-ACLF M HBV-ACLF M HBV-ACLF F HBV-ACLF F
5 HBV-ACLF M HBV-ACLF M NA HBV-ACLF M HBV-ACLF F HBV-ACLF M HBV-ACLF M HBV-ACLF M HBV-ACLF M HBV-ACLF M
6 Supplemental Table S3. List of the 2381 differentially expressed genes. Gene ID Gene short name Locus XLOC_ CICP27 1: XLOC_ ISG15 1: XLOC_ PRKCZ 1: XLOC_ SKI 1: XLOC_ PLCH2 1: XLOC_ VAMP3 1: XLOC_ NMNAT1,RP11-807G9.2 1: XLOC_ RBP7 1: XLOC_ PGD 1: XLOC_ UBIAD1 1: XLOC_ FBXO44 1: XLOC_ DRAXIN 1: XLOC_ TNFRSF8 1: XLOC_ MIR7846,TNFRSF1B 1: XLOC_ PADI4 1: XLOC_ OTUD3 1: XLOC_ CDA 1: XLOC_ NBPF3 1: XLOC_ ALPL 1: XLOC_ ZBTB40 1: XLOC_ RCAN3 1: XLOC_ CLIC4 1: XLOC_ : XLOC_ LDLRAP1 1: XLOC_ MAN1C1 1: XLOC_ SYTL1 1: XLOC_ FGR 1: XLOC_ RNU6-1245P,STX12 1: XLOC_ THEMIS2 1: XLOC_ : XLOC_ EPB41 1: XLOC_ SERINC2 1: XLOC_ LCK 1: XLOC_ AGO4 1: XLOC_ MIR6732,ZC3H12A 1: XLOC_ EXO5 1: XLOC_ PPIH,RP5-994D16.3 1: XLOC_ BTBD19,PLK3 1: XLOC_ FAAHP1 1: XLOC_ GPX7 1: XLOC_ PODN 1: XLOC_ C1orf177,DHCR24 1: XLOC_ LEPR,LEPROT 1: XLOC_ PDE4B 1: XLOC_ TCTEX1D1 1: XLOC_ IL12RB2 1: XLOC_ GADD45A 1: XLOC_ PTGFR 1:
7 XLOC_ PRKACB 1: XLOC_ CDC14A 1: XLOC_ RP4-575N6.5,S1PR1 1: XLOC_ GPSM2 1: XLOC_ CELSR2 1: XLOC_ CYB561D1 1: XLOC_ GSTM1,GSTM2,GSTM4 1: XLOC_ SLC22A15 1: XLOC_ CD2 1: XLOC_ LINC : XLOC_ FCGR1B 1: XLOC_ FCGR1C,RP5-998N21.4 1: XLOC_ LINC00869,PPIAL4C,RP11-353N4.6 1: XLOC_ FCGR1A 1: XLOC_ SNX27 1: XLOC_ AL : XLOC_ S100A9 1: XLOC_ ATP8B2 1: XLOC_ IL6R 1: XLOC_ ZBTB7B 1: XLOC_ SEMA4A 1: XLOC_ RP11-71G12.1 1: XLOC_ : XLOC_ CD1D 1: XLOC_ MNDA 1: XLOC_ FCER1A 1: XLOC_ FCRL6 1: XLOC_ SLAMF7 1: XLOC_ FCER1G,MIR5187,NDUFS2,TOMM 40L 1: XLOC_ FCGR2A,FCGR2B,FCGR2C,HSPA6,HSPA7,RP11-25K21.6 1: XLOC_ ATF6 1: XLOC_ C1orf226,NOS1AP,RP11-565P22.6 1: XLOC_ UAP1 1: XLOC_ CD247 1: XLOC_ XCL1 1: XLOC_ FASLG 1: XLOC_ QSOX1 1: XLOC_ NPL 1: XLOC_ LAMC1 1: XLOC_ RGL1 1: XLOC_ C1orf21 1: XLOC_ PLA2G4A 1: XLOC_ RGS18 1: XLOC_ RGS1 1: XLOC_ RGS2 1: XLOC_ GPR25 1: XLOC_ C1orf106 1: XLOC_ LGR6 1:
8 XLOC_ PPFIA4 1: XLOC_ BTG2 1: XLOC_ ATP2B4 1: XLOC_ LAX1 1: XLOC_ SOX13 1: XLOC_ SRGAP2 1: XLOC_ PFKFB2 1: XLOC_ CD55 1: XLOC_ CR1 1: XLOC_ G0S2 1: XLOC_ TRAF3IP3 1: XLOC_ ATF3 1: XLOC_ Mar 1: XLOC_ HLX 1: XLOC_ DEGS1 1: XLOC_ H3F3A 1: XLOC_ ARF1,MIR3620 1: XLOC_ OBSCN 1: XLOC_ FTH1P2 1: XLOC_ RHOU 1: XLOC_ CNST 1: XLOC_ SCCPDH 1: XLOC_ NLRP3 1: XLOC_ LYPD8 1: XLOC_ HES4 1: XLOC_ GPR153 1: XLOC_ PLEKHG5,TNFRSF25 1: XLOC_ SLC2A5 1: XLOC_ DHRS3 1: XLOC_ ARHGEF19 1: XLOC_ PADI2 1: XLOC_ PADI4 1: XLOC_ ALDH4A1,IFFO2,RP13-279N23.2,T AS1R2 1: XLOC_ ECE1 1: XLOC_ NBPF2P 1: XLOC_ TCEA3 1: XLOC_ E2F2 1: XLOC_ RUNX3 1: XLOC_ : XLOC_ PAFAH2 1: XLOC_ ZNF683 1: XLOC_ AHDC1 1: XLOC_ FGR 1: XLOC_ LAPTM5 1: XLOC_ SDC3 1: XLOC_ LCK 1: XLOC_ RNF19B 1: XLOC_ A3GALT2,PHC2 1: XLOC_ CLSPN 1:
9 XLOC_ COL8A2 1: XLOC_ CSF3R,MRPS15 1: XLOC_ MTF1,YRDC 1: XLOC_ HPCAL4 1: XLOC_ RIMS3 1: XLOC_ CITED4 1: XLOC_ : XLOC_ SLC2A1 1: XLOC_ SPATA6 1: XLOC_ ORC1 1: XLOC_ ECHDC2 1: XLOC_ SLC1A7 1: XLOC_ SSBP3 1: XLOC_ TTC22 1: XLOC_ JUN 1: XLOC_ JAK1 1: XLOC_ GNG5 1: XLOC_ CTBS 1: XLOC_ MIR7856,ODF2L 1: XLOC_ GBP5 1: XLOC_ TGFBR3 1: XLOC_ EVI5 1: XLOC_ ARHGAP29 1: XLOC_ RPL7P9 1: XLOC_ DPYD 1: XLOC_ SLC25A24 1: XLOC_ PSRC1 1: XLOC_ SORT1 1: XLOC_ AMIGO1 1: XLOC_ KCNA3 1: XLOC_ PPM1J,RHOC,RP11-426L : XLOC_ BCL2L15 1: XLOC_ CD2 1: XLOC_ FTH1P22 1: XLOC_ ZNF697 1: XLOC_ NOTCH2,RP5-1042I8.7 1: XLOC_ OTUD7B 1: XLOC_ CTSS 1: XLOC_ LINGO4,RORC 1: XLOC_ THEM4 1: XLOC_ S100A11 1: XLOC_ IVL 1: XLOC_ S100A12 1: XLOC_ S100A8 1: XLOC_ S100A6 1: XLOC_ RAB13 1: XLOC_ PBXIP1 1: XLOC_ GBA 1: XLOC_ RIT1 1: XLOC_ IQGAP3 1:
10 XLOC_ ARHGEF11 1: XLOC_ FCRL5 1: XLOC_ FCRL3 1: XLOC_ FCRL6 1: XLOC_ IGSF8 1: XLOC_ SLAMF6 1: XLOC_ SLAMF1 1: XLOC_ SH2D1B 1: XLOC_ GPA33 1: XLOC_ CD247 1: XLOC_ CREG1 1: XLOC_ XCL2 1: XLOC_ F5 1: XLOC_ KIFAP3 1: XLOC_ RP11-568K15.1,RP4-798P15.3,SEC1 6B 1: XLOC_ GLUL 1: XLOC_ NCF2 1: XLOC_ ARPC5 1: XLOC_ COLGALT2 1: XLOC_ EDEM3 1: XLOC_ PTGS2 1: XLOC_ : XLOC_ KIF21B 1: XLOC_ ARL8A 1: XLOC_ ADIPOR1 1: XLOC_ CHI3L1 1: XLOC_ CHIT1 1: XLOC_ ETNK2 1: XLOC_ RAB29 1: XLOC_ SLC41A1 1: XLOC_ IL10 1: XLOC_ FAIM3 1: XLOC_ PLXNA2 1: XLOC_ SLC30A1 1: XLOC_ LPGAT1 1: XLOC_ TLR5 1: XLOC_ LBR 1: XLOC_ RP11-285F7.2,TMEM63A 1: XLOC_ PARP1 1: XLOC_ ITPKB 1: XLOC_ : XLOC_ PCNXL2,RP5-862P8.3 1: XLOC_ LYST 1: XLOC_ CHRM3-AS2 1: XLOC_ CEP170 1: XLOC_ ZNF692 1: XLOC_ : XLOC_ : XLOC_ :
11 XLOC_ ZMYND11 10: XLOC_ AKR1C1,AKR1C3,RP11-499O7.4 10: XLOC_ PFKFB3 10: XLOC_ PRKCQ-AS1,RP11-554I8.1 10: XLOC_ GATA3 10: XLOC_ ECHDC3 10: XLOC_ OPTN 10: XLOC_ VIM 10: XLOC_ PLXDC2 10: XLOC_ MSRB2 10: XLOC_ OTUD1 10: XLOC_ APBB1IP 10: XLOC_ MAP3K8 10: XLOC_ HNRNPA1P32 10: XLOC_ ZEB1 10: XLOC_ CSGALNACT2 10: XLOC_ ALOX5 10: XLOC_ : XLOC_ ASAH2B 10: XLOC_ UBE2D1 10: XLOC_ CDK1 10: XLOC_ ZNF365 10: XLOC_ : XLOC_ SRGN 10: XLOC_ HKDC1 10: XLOC_ TSPAN15 10: XLOC_ C10orf35 10: XLOC_ COL13A1 10: XLOC_ DDIT4 10: XLOC_ MCU 10: XLOC_ PLAU 10: XLOC_ ADK,MRPL35P3 10: XLOC_ KAT6B 10: XLOC_ SAMD8 10: XLOC_ C10orf11 10: XLOC_ PPIF 10: XLOC_ CDHR1 10: XLOC_ CCSER2 10: XLOC_ LIPN 10: XLOC_ CEP55 10: XLOC_ TBC1D12 10: XLOC_ C10orf131,CC2D2B,ENTPD1,RP11-248J23.6,RP11-248J23.7,RP11-429G 10: ,RP11-690P14.4 XLOC_ FRAT1 10: XLOC_ GOLGA7B 10: XLOC_ SCD 10: XLOC_ ELOVL3 10: XLOC_ GSTO1,GSTO2 10: XLOC_ SHOC2 10:
12 XLOC_ ENO4 10: XLOC_ GRK5 10: XLOC_ PLEKHA1 10: XLOC_ PTPRE,RP11-4C : XLOC_ VENTX 10: XLOC_ CYP2E1,MTG1,PAOX,RP11-108K1 4.8,SCART1 10: XLOC_ DIP2C 10: XLOC_ GDI2 10: XLOC_ PRKCQ 10: XLOC_ SFMBT2 10: XLOC_ NMT2 10: XLOC_ FAM171A1 10: XLOC_ ST8SIA6 10: XLOC_ MPP7 10: XLOC_ SVILP1 10: XLOC_ KIF5B 10: XLOC_ NAMPTP1 10: XLOC_ RP11-258F22.1,ZNF248 10: XLOC_ NCOA4 10: XLOC_ ANTXRLP1 10: XLOC_ C10orf128 10: XLOC_ SGMS1 10: XLOC_ IPMK 10: XLOC_ CCDC6 10: XLOC_ ANK3 10: XLOC_ RTKN2 10: XLOC_ EGR2 10: XLOC_ AIFM2,TYSND1 10: XLOC_ PRF1 10: XLOC_ PSAP 10: XLOC_ SPOCK2 10: XLOC_ MICU1 10: XLOC_ ZNF503 10: XLOC_ DLG5 10: XLOC_ ANKRD22 10: XLOC_ CPEB3 10: XLOC_ MYOF 10: XLOC_ ALDH18A1 10: XLOC_ PIK3AP1 10: XLOC_ FRAT2 10: XLOC_ DNMBP 10: XLOC_ CHUK,ERLIN1 10: XLOC_ FBXW4 10: XLOC_ NT5C2 10: XLOC_ COL17A1,MIR936 10: XLOC_ ITPRIP 10: XLOC_ ABLIM1 10: XLOC_ ENO4,KIAA : XLOC_ PRDX3 10:
13 XLOC_ CHST15 10: XLOC_ MKI67 10: XLOC_ : XLOC_ : XLOC_ : XLOC_ : XLOC_ ATHL1,RP11-326C3.2 11: XLOC_ IFITM1,IFITM2,RP11-326C3.10,RP C3.11,RP11-326C3.12,RP : C3.15 XLOC_ SIGIRR 11: XLOC_ RASSF7,RP11-496I9.1 11: XLOC_ TALDO1 11: XLOC_ AC ,LSP1,hsa-mir : XLOC_ TNNT3 11: XLOC_ CD81 11: XLOC_ OR52L2P 11: XLOC_ GVINP1 11: XLOC_ SWAP70 11: XLOC_ ADM 11: XLOC_ AMPD3 11: XLOC_ FAR1 11: XLOC_ LDHA 11: XLOC_ PRRG4 11: XLOC_ CAT 11: XLOC_ CD44 11: XLOC_ LDLRAD3 11: XLOC_ PRR5L 11: XLOC_ CD82 11: XLOC_ SPI1 11: XLOC_ PTPRJ 11: XLOC_ OR10Y1P,STX3 11: XLOC_ MS4A3 11: XLOC_ MS4A4A 11: XLOC_ MS4A14,MS4A6E,MS4A7 11: XLOC_ CD6 11: XLOC_ CD5 11: XLOC_ LGALS12 11: XLOC_ RARRES3 11: XLOC_ RTN3 11: XLOC_ NAA40 11: XLOC_ SLC22A20 11: XLOC_ CTSW 11: XLOC_ RAD9A,TBC1D10C 11: XLOC_ ALDH3B1 11: XLOC_ FOLR3 11: XLOC_ INPPL1 11: XLOC_ P2RY2,RP11-800A3.4 11: XLOC_ RELT,RP11-809N8.4 11: XLOC_ PLEKHB1 11:
14 XLOC_ RPS3,SNORD15A 11: XLOC_ SERPINH1 11: XLOC_ DGAT2 11: XLOC_ ACER3 11: XLOC_ CAPN5 11: XLOC_ MYO7A 11: XLOC_ RPS28P7 11: XLOC_ : XLOC_ PRSS23 11: XLOC_ SLC36A4 11: XLOC_ FUT4,PIWIL4,RP11-867G2.4 11: XLOC_ RP11-60C6.5,SRSF8 11: XLOC_ ENDOD1 11: XLOC_ RAB39A 11: XLOC_ ZC3H12C 11: XLOC_ NCAM1 11: XLOC_ ZBTB16 11: XLOC_ AP ,PAFAH1B2,SIDT2,TA GLN 11: XLOC_ RP11-728F : XLOC_ : XLOC_ CD3E 11: XLOC_ CD3G 11: XLOC_ TMEM25,TTC36 11: XLOC_ VWA5A 11: XLOC_ ROBO3 11: XLOC_ SLC37A2 11: XLOC_ APLP2 11: XLOC_ ST14 11: XLOC_ GLB1L2 11: XLOC_ ANO9,SIGIRR 11: XLOC_ CTSD,IFITM10,KRTAP5-4,RP K3.1 11: XLOC_ CDKN1C 11: XLOC_ OSBPL5 11: XLOC_ RHOG 11: XLOC_ PRKCDBP 11: XLOC_ APBB1 11: XLOC_ DCHS1 11: XLOC_ CYB5R2 11: XLOC_ DENND5A 11: XLOC_ RP11-1H15.1,SBF2 11: XLOC_ RP11-351I : XLOC_ MTRNR2L8,RNF141 11: XLOC_ MRVI1 11: XLOC_ DKK3 11: XLOC_ RP11-21L19.1,RRAS2 11: XLOC_ E2F8 11: XLOC_ LMO2 11: XLOC_ TP53I11 11:
15 XLOC_ MYBPC3 11: XLOC_ RP11-750H9.5,SPI1 11: XLOC_ PRG2,RP11-872D17.8,SLC43A3 11: XLOC_ SLC43A1 11: XLOC_ MPEG1 11: XLOC_ TCN1 11: XLOC_ AP ,MS4A6A 11: XLOC_ MIR6503,MS4A4E 11: XLOC_ FTH1 11: XLOC_ MEN1 11: XLOC_ CDC42BPG 11: XLOC_ LTBP3 11: XLOC_ AP5B1 11: XLOC_ FOSL1 11: XLOC_ CD248 11: XLOC_ SLC29A2 11: XLOC_ CTSF 11: XLOC_ CORO1B,PTPRCAP 11: XLOC_ UNC93B1,UNC93B5 11: XLOC_ CPT1A 11: XLOC_ KCNE3 11: XLOC_ PAK1 11: XLOC_ GAB2 11: XLOC_ PRCP 11: XLOC_ SYTL2 11: XLOC_ PICALM 11: XLOC_ ME3 11: XLOC_ SLC36A4 11: XLOC_ SMCO4 11: XLOC_ RP11-680H : XLOC_ MMP8 11: XLOC_ PDGFD 11: XLOC_ RP11-693N9.2 11: XLOC_ CARD16,CARD17,CASP1 11: XLOC_ KDELC2 11: XLOC_ USP28 11: XLOC_ RP11-109L : XLOC_ FXYD2,FXYD6,FXYD6-FXYD2 11: XLOC_ CD3D 11: XLOC_ BCL9L 11: XLOC_ HYOU1 11: XLOC_ HSPA8,SNORD14C,SNORD14D 11: XLOC_ FEZ1 11: XLOC_ : XLOC_ ETS1 11: XLOC_ B3GAT1 11: XLOC_ : XLOC_ : XLOC_ CCND2 12: XLOC_ LTBR,RP1-102E :
16 XLOC_ GAPDH 12: XLOC_ LAG3 12: XLOC_ C12orf57,RNU7-1 12: XLOC_ CLEC4A,FAM66C,ZNF705A 12: XLOC_ CLEC6A 12: XLOC_ CLEC4D 12: XLOC_ A2MP1,KRT17P8,LINC00987,RP11-118B22.4,RP11-436I9.6 12: XLOC_ RP11-726G1.1 12: XLOC_ CLEC2D,GOT2P3,RP11-705C15.2,R P11-705C15.3,RP11-705C15.5,RP11-12: L1.1 XLOC_ KLRF1 12: XLOC_ CLEC7A 12: XLOC_ TMEM52B 12: XLOC_ KLRD1 12: XLOC_ RP11-277P : XLOC_ RP11-291B : XLOC_ ETV6 12: XLOC_ HTR7P1,RP11-377D9.3 12: XLOC_ PTPRO 12: XLOC_ MGST1 12: XLOC_ PLEKHA5 12: XLOC_ SSPN 12: XLOC_ FAR2 12: XLOC_ FGD4 12: XLOC_ LRRK2 12: XLOC_ METTL7A 12: XLOC_ ACVRL1 12: XLOC_ ACVR1B 12: XLOC_ GRASP 12: XLOC_ NR4A1 12: XLOC_ SOAT2 12: XLOC_ ESPL1 12: XLOC_ SP1 12: XLOC_ CDK2,DGKA 12: XLOC_ COQ10A 12: XLOC_ LRP1 12: XLOC_ DTX3 12: XLOC_ ARHGEF25 12: XLOC_ MSRB3 12: XLOC_ IRAK3,MIR : XLOC_ DYRK2 12: XLOC_ LYZ 12: XLOC_ : XLOC_ ATXN7L3B 12: XLOC_ GLIPR1 12: XLOC_ TMTC2 12: XLOC_ SOCS2 12: XLOC_ PLXNC1 12:
17 XLOC_ APAF1 12: XLOC_ DRAM1 12: XLOC_ KIAA : XLOC_ TMEM263 12: XLOC_ UNG 12: XLOC_ SH2B3 12: XLOC_ ACAD10,ALDH2,RP11-162P : XLOC_ RPH3A 12: XLOC_ SDSL 12: XLOC_ CCDC64 12: XLOC_ P2RX7,RP11-340F : XLOC_ CAMKK2 12: XLOC_ ORAI1 12: XLOC_ TCTN2 12: XLOC_ GLT1D1 12: XLOC_ FBXL14 12: XLOC_ TNFRSF1A 12: XLOC_ LPAR5 12: XLOC_ C1R,C1RL 12: XLOC_ CD163,CD163L1 12: XLOC_ CLEC4C 12: XLOC_ SLC2A3 12: XLOC_ C3AR1 12: XLOC_ CLEC4E,RP11-561P : XLOC_ KLRB1 12: XLOC_ CLEC7A 12: XLOC_ OLR1 12: XLOC_ KLRC1,KLRC2,KLRC3,KLRC4,KL RC4-KLRK1,KLRK1,NKG2-E,RP11 12: P12.9 XLOC_ STYK1 12: XLOC_ YBX3 12: XLOC_ MANSC1 12: XLOC_ PLBD1 12: XLOC_ LDHB 12: XLOC_ BCAT1 12: XLOC_ KRAS 12: XLOC_ TMTC1 12: XLOC_ PKP2 12: XLOC_ CPNE8 12: XLOC_ ABCD2,AC ,KIF21A 12: XLOC_ NELL2 12: XLOC_ SLC38A1 12: XLOC_ PCED1B-AS1 12: XLOC_ VDR 12: XLOC_ WNT10B 12: XLOC_ RHEBL1 12: XLOC_ NCKAP5L 12: XLOC_ LIMA1 12: XLOC_ KRT72,KRT73 12:
18 XLOC_ ITGB7 12: XLOC_ GPR84 12: XLOC_ TESPA1 12: XLOC_ CD63 12: XLOC_ AGAP2 12: XLOC_ GNS 12: XLOC_ AC ,RBMS1P1,RP11-335I : XLOC_ RP11-81H14.1,RP11-81H : XLOC_ LIN7A 12: XLOC_ DUSP6 12: XLOC_ RP11-887P2.3 12: XLOC_ NTN4 12: XLOC_ LTA4H 12: XLOC_ SLC9A7P1 12: XLOC_ RP11-135F9.3,UHRF1BP1L 12: XLOC_ NT5DC3,RP11-341G : XLOC_ CKAP4 12: XLOC_ MTERF2 12: XLOC_ CRY1 12: XLOC_ CMKLR1 12: XLOC_ CORO1C 12: XLOC_ TRPV4 12: XLOC_ GLTP 12: XLOC_ ANAPC7,ARPC3,RP11-478C : XLOC_ HVCN1 12: XLOC_ TESC 12: XLOC_ WSB2 12: XLOC_ GCN1L1 12: XLOC_ CAMKK2 12: XLOC_ CLIP1 12: XLOC_ RILPL2 12: XLOC_ LINC : XLOC_ RNU6-1017P 12: XLOC_ : XLOC_ C1QTNF9,RP11-307N16.6,SPATA1 3 13: XLOC_ PABPC3 13: XLOC_ NUPL1 13: XLOC_ ALOX5AP 13: XLOC_ KL 13: XLOC_ LACC1 13: XLOC_ ARL11 13: XLOC_ HNRNPA1L2,MRPS31P4,RP11-78J : XLOC_ OLFM4 13: XLOC_ KLF5 13: XLOC_ LINC00381,LINC00402,RPL21P108 13: XLOC_ AC : XLOC_ DNAJC3 13:
19 XLOC_ TM9SF2 13: XLOC_ RP11-153I24.4,TNFSF13B 13: XLOC_ IRS2 13: XLOC_ ATP11A 13: XLOC_ MCF2L 13: XLOC_ TMCO3 13: XLOC_ : XLOC_ ZDHHC20 13: XLOC_ SACS 13: XLOC_ MTMR6 13: XLOC_ FLT3 13: XLOC_ SLC7A1 13: XLOC_ LCP1 13: XLOC_ KIAA0226L,LINC00563,PPP1R2P4 13: XLOC_ DLEU2,MIR15A,MIR : XLOC_ PCDH9 13: XLOC_ DACH1 13: XLOC_ KLF12 13: XLOC_ TBC1D4 13: XLOC_ AC ,KCTD12 13: XLOC_ DOCK9 13: XLOC_ CCR12P 13: XLOC_ IRS2 13: XLOC_ RAB20 13: XLOC_ CARS2 13: XLOC_ LINC00565,RASA3 13: XLOC_ : XLOC_ : XLOC_ : XLOC_ RP11-203M5.6 14: XLOC_ ANG,RNASE4 14: XLOC_ RNASE6,RP11-219E7.1 14: XLOC_ RNASE3 14: XLOC_ RP11-84C : XLOC_ RNASE2 14: XLOC_ ARHGEF40 14: XLOC_ TRAV1-1 14: XLOC_ TRAV8-2,TRAV8-3 14: XLOC_ TRAV : XLOC_ TRAV : XLOC_ TRAV : XLOC_ TRAV9-2 14: XLOC_ TRAV : XLOC_ TRAV16 14: XLOC_ TRAV17 14: XLOC_ TRAV19 14: XLOC_ TRDV1 14: XLOC_ TRAV24 14: XLOC_ TRAV25 14: XLOC_ TRAV :
20 XLOC_ TRAV27 14: XLOC_ TRAV29DV5 14: XLOC_ AE ,TRAC,TRAJ21,TRAJ2 3,TRAJ29,TRAJ32,TRAJ39,TRAJ54, TRAV38-2DV8,TRDC,TRDD3,TRD 14: J1,TRDJ2,TRDJ3,TRDJ4,TRDV2 XLOC_ RP11-104E : XLOC_ LGALS3 14: XLOC_ PELI2 14: XLOC_ FLJ22447,PRKCH,RP11-47I : XLOC_ HIF1A,RP11-618G20.1,SNAPC1 14: XLOC_ SYNE2 14: XLOC_ PLEKHG3 14: XLOC_ SIPA1L1 14: XLOC_ PSEN1 14: XLOC_ FOS 14: XLOC_ JDP2 14: XLOC_ FLVCR2,IFT43,TTLL5 14: XLOC_ CTD-2341M : XLOC_ SLC24A4 14: XLOC_ CTD-2506J : XLOC_ RP11-204N : XLOC_ EVL 14: XLOC_ : XLOC_ RCOR1 14: XLOC_ TDRD9 14: XLOC_ CRIP2 14: XLOC_ TMEM121 14: XLOC_ MIR6717,NDRG2 14: XLOC_ SALL2 14: XLOC_ OR10G2 14: XLOC_ SLC7A7 14: XLOC_ AJUBA,HAUS4,MIR4707,RP I3.5 14: XLOC_ CDH24 14: XLOC_ CEBPE 14: XLOC_ CTSG 14: XLOC_ GZMH 14: XLOC_ GZMB 14: XLOC_ SPTSSA 14: XLOC_ BAZ1A 14: XLOC_ NFKBIA 14: XLOC_ RPL32P29,RPS29 14: XLOC_ PYGL 14: XLOC_ ERO1L 14: XLOC_ CNIH1 14: XLOC_ GMFB 14: XLOC_ DLGAP5 14: XLOC_ WDR89 14: XLOC_ ZFP36L1 14:
21 XLOC_ ACTN1,HMGN1P3 14: XLOC_ NUMB,RP1-240K6.3,RP4-647C : XLOC_ PNMA1 14: XLOC_ LTBP2,MIR4709,NPC2 14: XLOC_ TMED8 14: XLOC_ SPTLC2 14: XLOC_ GPR68 14: XLOC_ CCDC88C 14: XLOC_ CATSPERB,TC2N 14: XLOC_ FBLN5 14: XLOC_ DDX24 14: XLOC_ SERPINA1 14: XLOC_ CLMN,CTD-2240H : XLOC_ LINC : XLOC_ BCL11B 14: XLOC_ : XLOC_ WARS 14: XLOC_ CDC42BPB 14: XLOC_ CKB 14: XLOC_ PPP1R13B 14: XLOC_ : XLOC_ IGHV : XLOC_ : XLOC_ : XLOC_ : XLOC_ RP11-603B : XLOC_ WHAMMP3 15: XLOC_ NIPA1,RP11-566K : XLOC_ MKRN3 15: XLOC_ PWAR5,PWAR6,RP11-701H24.7,SN HG14,SNORD107,SNORD115-13,S NORD115-26,SNORD115-46,SNOR D115-7,SNORD116,SNORD116-20, 15: SNORD116-22,SNORD116-3,SNOR D116-4,SNORD116-5,SNORD64,SN RPN,SNURF XLOC_ WHAMMP2 15: XLOC_ APBA2 15: XLOC_ HMGN2P5 15: XLOC_ GOLGA8B 15: XLOC_ THBS1 15: XLOC_ BAHD1 15: XLOC_ CASC5 15: XLOC_ SPINT1 15: XLOC_ CHP1 15: XLOC_ NUSAP1 15: XLOC_ BLOC1S6,HMGN2P46,RP11-96O20.4,SQRDL 15: XLOC_ AQP9 15: XLOC_ RPS3AP6 15:
22 XLOC_ LACTB 15: XLOC_ USP3 15: XLOC_ AC ,ANKDD1A,PLEKHO2 15: XLOC_ SMAD3 15: XLOC_ KIF23 15: XLOC_ SCAMP5 15: XLOC_ C15orf39 15: XLOC_ ST20-AS1 15: XLOC_ BCL2A1 15: XLOC_ IL16 15: XLOC_ ABHD2 15: XLOC_ LINC : XLOC_ ANPEP 15: XLOC_ IQGAP1 15: XLOC_ FURIN 15: XLOC_ FES,MAN2A2 15: XLOC_ SLCO3A1 15: XLOC_ ARRDC4 15: XLOC_ CYFIP1 15: XLOC_ ATP10A 15: XLOC_ LPCAT4 15: XLOC_ GOLGA8A 15: XLOC_ RASGRP1 15: XLOC_ CTD-2033D15.1,CTD-2033D : XLOC_ BMF 15: XLOC_ LTK 15: XLOC_ FBN1 15: XLOC_ ATP8B4 15: XLOC_ DMXL2 15: XLOC_ GNB5 15: XLOC_ PIGBOS1,RAB27A 15: XLOC_ CCPG1,DYX1C1,DYX1C1-CCPG1, MIR628 15: XLOC_ NEDD4 15: XLOC_ ALDH1A2 15: XLOC_ AQP9 15: XLOC_ BNIP2 15: XLOC_ : XLOC_ ANXA2 15: XLOC_ RORA,uc_338 15: XLOC_ : XLOC_ DAPK2 15: XLOC_ FAM96A 15: XLOC_ OAZ2 15: XLOC_ UACA 15: XLOC_ PKM 15: XLOC_ ADPGK 15: XLOC_ NPTN 15: XLOC_ IMP3 15: XLOC_ CTSH 15:
23 XLOC_ RASGRF1 15: XLOC_ BCL2A1 15: XLOC_ AC ,GOLGA2P10 15: XLOC_ SEC11A 15: XLOC_ HAPLN3 15: XLOC_ MFGE8 15: XLOC_ ANPEP 15: XLOC_ CHSY1 15: XLOC_ TARSL2 15: XLOC_ : XLOC_ HBA2 16: XLOC_ CACNA1H 16: XLOC_ TMEM204 16: XLOC_ LA16c-380H5.5 16: XLOC_ MMP25 16: XLOC_ IL32,RNU1-125P,RNU1-22P 16: XLOC_ HMOX2 16: XLOC_ C16orf72,RP11-473I1.9 16: XLOC_ CTD-2349B8.1,ITPRIPL2,SYT17 16: XLOC_ METTL9 16: XLOC_ PRKCB 16: XLOC_ IL4R 16: XLOC_ IL21R 16: XLOC_ SBK1 16: XLOC_ APOBR 16: XLOC_ LAT,RP11-264B17.3,SPNS1 16: XLOC_ AC ,QPRT,SPN 16: XLOC_ ALDOA 16: XLOC_ ITGAL 16: XLOC_ ITGAM 16: XLOC_ ITGAX 16: XLOC_ RBL2 16: XLOC_ LPCAT2,RP11-212I : XLOC_ CPNE2 16: XLOC_ GPR114,HMGB3P32 16: XLOC_ GPR56 16: XLOC_ GPR97 16: XLOC_ NDRG4 16: XLOC_ LINC : XLOC_ PDP2,RP11-61A14.2,RP11-61A : XLOC_ NOL3 16: XLOC_ RLTPR 16: XLOC_ PLA2G15 16: XLOC_ HP,HPR 16: XLOC_ ATP2C2 16: XLOC_ CRISPLD2 16: XLOC_ KIAA : XLOC_ ZFPM1 16: XLOC_ : XLOC_ CDT1 16:
24 XLOC_ VPS9D1-AS1,ZNF276 16: XLOC_ AC ,FAM157C 16: XLOC_ AXIN1 16: XLOC_ RAB11FIP3 16: XLOC_ MSRB1 16: XLOC_ ABCA3 16: XLOC_ PRSS30P 16: XLOC_ PKMYT1 16: XLOC_ RP11-473M : XLOC_ MEFV 16: XLOC_ NLRC3 16: XLOC_ RP11-461A8.4,TRAP1 16: XLOC_ CDIP1 16: XLOC_ CPPED1 16: XLOC_ IGSF6 16: XLOC_ SBK1 16: XLOC_ RP11-297C4.6,SEPT1 16: XLOC_ PYCARD 16: XLOC_ HMGN2P41 16: XLOC_ RP11-44F14.1,RP11-44F : XLOC_ CES1 16: XLOC_ CCDC102A 16: XLOC_ GPR114 16: XLOC_ GPR56 16: XLOC_ NAE1 16: XLOC_ ATP6V0D1 16: XLOC_ MLKL 16: XLOC_ CDYL2 16: XLOC_ COTL1 16: XLOC_ KLHDC4,RP11-278A : XLOC_ P2RX1 17: XLOC_ SPNS2 17: XLOC_ ARRB2 17: XLOC_ ASGR2 17: XLOC_ RPL7AP64 17: XLOC_ NLGN2 17: XLOC_ CD68,EIF4A1,MPDU1,SENP3,SEN P3-EIF4A1,SNORA48,SNORA67,S NORD10,TNFSF12,TNFSF12-TNFS 17: F13,TNFSF13 XLOC_ KDM6B,TMEM88 17: XLOC_ CHD3 17: XLOC_ HS3ST3B1 17: XLOC_ CCDC144A,NOS2P4,RP11-219A15. 1,RP11-219A15.2,RP11-219A15.4,U 17: SP32P1 XLOC_ KRT16P4,LGALS9C 17: XLOC_ CCDC144CP,SPECC1,USP32P3 17: XLOC_ RP11-173M1.7,RP11-173M1.8,WSB 1 17:
25 XLOC_ CPD 17: XLOC_ ADAP2,RN7SL138P,RNF135,RP11-848P1.3 17: XLOC_ RHOT1 17: XLOC_ SLFN5 17: XLOC_ AC ,RDM1 17: XLOC_ DUSP14 17: XLOC_ CISD3,CTB-58E17.1,MLLT6 17: XLOC_ ERBB2 17: XLOC_ IKZF3 17: XLOC_ THRA 17: XLOC_ RARA 17: XLOC_ : XLOC_ ATP6V0A1,MIR5010,RP11-400F : XLOC_ GRN 17: XLOC_ TBX21 17: XLOC_ ABCC3 17: XLOC_ PCTP 17: XLOC_ NOG 17: XLOC_ SCPEP1 17: XLOC_ DYNLL2,RP11-159D : XLOC_ YPEL2 17: XLOC_ CLTC,VMP1 17: XLOC_ CA4 17: XLOC_ MILR1 17: XLOC_ RGS9 17: XLOC_ ARSG,PRKAR1A,RP11-120M : XLOC_ KCNJ2 17: XLOC_ RPL38 17: XLOC_ KIF19 17: XLOC_ MIR3615,RAB37,SLC9A3R1 17: XLOC_ LLGL2,TSEN54 17: XLOC_ SCARNA16,SEC14L1,SNHG20 17: XLOC_ SEPT9 17: XLOC_ TMC8 17: XLOC_ SYNGR2 17: XLOC_ BIRC5 17: XLOC_ GAA 17: XLOC_ RPTOR 17: XLOC_ MIR6787,RP13-516M14.10,RP M14.8,SLC16A3 17: XLOC_ RP E : XLOC_ DOC2B 17: XLOC_ FAM101B 17: XLOC_ YWHAE 17: XLOC_ CRK 17: XLOC_ SLC43A2 17: XLOC_ RILP,SCARF1 17: XLOC_ MIR22,MIR22HG 17:
26 XLOC_ RP11-147K16.3,SPATA22,TRPV3 17: XLOC_ CAMKK1,P2RX1 17: XLOC_ PELP1 17: XLOC_ CXCL16,RP11-314A : XLOC_ SCIMP 17: XLOC_ ASGR2,CLEC10A 17: XLOC_ ZBTB4 17: XLOC_ AURKB 17: XLOC_ GAS7 17: XLOC_ PMP22 17: XLOC_ TBC1D27,TNFRSF13B 17: XLOC_ CCDC144B,CTD-2303H24.2,FAM1 06A,USP32P2 17: XLOC_ FOXO3B,RP11-815I9.3,RP11-815I9. 4,TBC1D28,ZNF286B 17: XLOC_ RAB34 17: XLOC_ CTD-2370N5.3,EVI2A,EVI2B 17: XLOC_ RP M14.5,RP11-686D22.10,S LFN12L,SLFN13,TOMM20P2 17: XLOC_ HEATR9 17: XLOC_ CCL5 17: XLOC_ SYNRG 17: XLOC_ PLXDC1 17: XLOC_ IKZF3,RP11-94L : XLOC_ GSDMB,ORMDL3 17: XLOC_ TOP2A 17: XLOC_ CCR7 17: XLOC_ KRT23 17: XLOC_ JUP 17: XLOC_ LEPREL4 17: XLOC_ STAT3 17: XLOC_ LINC : XLOC_ VAT1 17: XLOC_ BRCA1 17: XLOC_ DUSP3 17: XLOC_ WNT3 17: XLOC_ TBX21 17: XLOC_ OSBPL7 17: XLOC_ SKAP1 17: XLOC_ LRRC59 17: XLOC_ MIR3614,TRIM25 17: XLOC_ CUEDC1 17: XLOC_ MPO 17: XLOC_ CYB561,RP11-269G : XLOC_ ICAM2 17: XLOC_ TEX2 17: XLOC_ PECAM1 17: XLOC_ AXIN2,CEP112,CTD-2535L : XLOC_ FAM20A 17: XLOC_ ABCA6,ABCA9 17:
27 XLOC_ CDC42EP4 17: XLOC_ CD300LB 17: XLOC_ CD300C 17: XLOC_ GRB2 17: XLOC_ TRIM65 17: XLOC_ ACOX1 17: XLOC_ SOCS3 17: XLOC_ CTD-2357A8.3 17: XLOC_ CYTH1 17: XLOC_ USP36 17: XLOC_ KIAA1731NL,TIMP2 17: XLOC_ BAIAP2-AS1 17: XLOC_ AATK 17: XLOC_ LINC : XLOC_ RP B8.4 17: XLOC_ : XLOC_ RAB40B 17: XLOC_ : XLOC_ : XLOC_ : XLOC_ EMILIN2,RP11-737O : XLOC_ RAB31 18: XLOC_ VAPA 18: XLOC_ IMPA2 18: XLOC_ ANKRD20A5P,CYP4F35P,RHOT1P 1,RP11-757O6.1,RP11-757O6.6 18: XLOC_ RIOK3 18: XLOC_ MAPRE2 18: XLOC_ SIGLEC15 18: XLOC_ CTIF,MIR : XLOC_ RAB27B 18: XLOC_ PMAIP1 18: XLOC_ SERPINB10,SERPINB2 18: XLOC_ HMSD,SERPINB8 18: XLOC_ TSHZ1 18: XLOC_ KCNG2 18: XLOC_ ENOSF1 18: XLOC_ YES1 18: XLOC_ ZBTB14 18: XLOC_ EPB41L3 18: XLOC_ RP11-973H7.1,RP11-973H7.4 18: XLOC_ RP11-535A5.1 18: XLOC_ DSC2 18: XLOC_ PSTPIP2 18: XLOC_ LOXHD1 18: XLOC_ SMAD7 18: XLOC_ MEX3C 18: XLOC_ BCL2 18: XLOC_ CD226,DOK6,RP11-543H : XLOC_ ZNF516 18:
28 XLOC_ GZMM 19: XLOC_ : XLOC_ FSTL3 19: XLOC_ AZU1 19: XLOC_ PRTN3 19: XLOC_ ELANE 19: XLOC_ ARID3A 19: XLOC_ MIDN 19: XLOC_ GADD45B 19: XLOC_ GNA15 19: XLOC_ AC ,TJP3 19: XLOC_ MATK 19: XLOC_ FSD1 19: XLOC_ SEMA6B 19: XLOC_ TNFSF9 19: XLOC_ EMR1 19: XLOC_ RETN 19: XLOC_ CTD-3214H19.16,MCEMP1,TRAPP C5 19: XLOC_ CTC-325H20.2,ZNF177,ZNF559,ZN F559-ZNF177 19: XLOC_ ICAM1 19: XLOC_ QTRT1 19: XLOC_ C19orf38 19: XLOC_ ZNF788 19: XLOC_ JUNB 19: XLOC_ CYP4F22 19: XLOC_ CYP4F3,CYP4F8 19: XLOC_ COLGALT1 19: XLOC_ RPL18A,SNORA68 19: XLOC_ IFI30,PIK3R2 19: XLOC_ LSM4 19: XLOC_ SLC25A42 19: XLOC_ PLEKHF1 19: XLOC_ URI1 19: XLOC_ ZNF507 19: XLOC_ DPY19L3 19: XLOC_ ZNF181 19: XLOC_ ZNF30 19: XLOC_ FFAR2 19: XLOC_ HAUS5 19: XLOC_ LRFN3 19: XLOC_ ZNF420 19: XLOC_ HKR1 19: XLOC_ ZFP36 19: XLOC_ LTBP4 19: XLOC_ AC : XLOC_ CEACAM3 19: XLOC_ ZNF574 19: XLOC_ CD177 19:
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
doi:10.1038/nature22977 Supplementary Data 1 Fig. 4d + CM (no MDK) + CM (MDK) 10 15 30 1h 12h 24h 10 15 30 1h 12h 24h P- P- Fig. 4e CM GoF CM LoF-M5 CM GoF MDK - + + - + + + Vehicle - + + - + + + Rapamycin
LAPTM4B is associated with poor prognosis in NSCLC and promotes the. NRF2-mediated stress response pathway in lung cancer cells
 LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells Yuho Maki, Junya Fujimoto, Wenhua Lang, Li Xu, Carmen Behrens, Ignacio I.
Supplementary Fig. 1. Nature Medicine: doi: /nm z Donor # BB-z. 28- IL2RB-z (YXXQ) 28-IL2RBz (YXXQ) Control MFI: 1866 MFI:
 Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Supplementary Fig. 1 a Donor # 28-IL2RB-z 28- IL2RB-z Control MFI: 1866 1394 612 1297 11 MFI: 1794 1294 566 1225 11 MFI: 1433 1157 327 1117 98.5 MFI: 1463 1224 352 111 126 MFI: 1441 1232 497 1268 151 MFI:
Nature Medicine doi: /nm.2772
 Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
Supplementary Table 1: FOXO3a and/or beta catenin regulated genes (12 h treatment) FC 4OHT FC DOX FC 4OHT+DOX FC 4OHT+DOX Known Gene Symbol vs. Vehicle P value* vs. Vehicle P value* vs. Vehicle P value*
TFDP2 TFDP1 SUMO1 SERTAD1 RPA3 RBBP8 RAD9A RAD17 RAD1 NBN MRE11A MNAT1 KPNA2 KNTC1 HUS1 HERC5 GTSE1 GTF2H1 GADD45A E2F4 DNM2 DDX11 CUL3 SKP2 RBL1 RB1
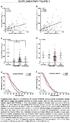
 -8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
-8-6 -4-2 CHEK2 MKI67 RB1 RBL1 SKP2 Fold-Change Fold-Change -3-2 -1 CCND1 CCNE1 CDK4 CDK5R1 5 1 15 ATM ATR BAX BCCIP BRCA1 BRCA2 BL1AT ABL1A ABL1 CCNG1 CCNG2 CCNH CCNT1 CCNT2 CDC2 CDC34 CDKN1A CDKN1B CDKN2A
Supplementary Methods
 Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Supplementary Methods Immunohistochemistry Five-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue specimens were deparaffinized and dehydrated. All sections were incubated with anti-human
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at
 Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Figure S1: Deletion of Dlx3 in the NC A) LacZ staining of Dlx3 LacZ/WT mice (a) and Wnt1- cre:r26r LacZ mice (b) at E11.5. The arrowhead points at Dlx3 expression in the branchial arches. B) PCR and Southern
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge.
 Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Supplementary Figure 1. Therapeutic efficacy of combined NDV and anti-ctla-4 therapy is attenuated with a larger tumor challenge. Bilateral flank B16-F10 tumors were established as described and the animals
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome
 Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Biological marks of early-life socioeconomic experience is detected in the adult inflammatory transcriptome Raphaële Castagné 1,2,3, Michelle Kelly-Irving 2,3, Gianluca Campanella 1, Florence Guida 1,
Lee et al., Suppl. Fig. 1
 Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Lee et al., Suppl. Fig. 1 Lee et al., Suppl. Fig. 2 Lee et al., Suppl. Fig. 3 Lee et al., Suppl. Fig. 4 Lee et al., Suppl. Fig. 5 Lee et al., Supplementary Table 1 Markers Percentage of p75+ cells [Mean
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications
 Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Table S1. Primary antibodies for Western blot analysis. Antibody name and specifications Manufacturer Actin (C-11) sc-1615, Santa Cruz, CA Akt1 (B-1) sc-5298 GAPDH (6C5) sc-32233 JAK2 (HR-758)
Supplementary Appendix; Figures S1-S16.
 Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Supplementary Appendix; Figures S1-S16. Supplementary Appendix; Figure S1. FACS-Purified reporter + T cells reveal greater transcriptional resolution than bulk CD4 cells. CD4 T cells were purified and
Nature Immunology: doi: /ni Supplementary Figure 1. IL-6 reporter mice.
 Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Supplementary Figure 1 IL-6 reporter mice. (a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this
Table S1. Full list of high-stringency HIF-1 binding sites
 Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Table S1. Full list of high-stringency HIF-1 HIF-1 ( Nearest gene locus 1 chr7 156986147 156987039 314 N DNAJB6 NM_058246 2 chr8 134456859 134457261 180 N NDRG1 NM_006096 3 chr1 114156539 114157002 288
Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC)
 Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
Additional file 1 Supplemental Table S1. Tumor specific networks are enriched with somatically mutated genes (taken from the database COSMIC) COSMIC genes in tumor network COSMIC genes not in tumor network
RARα2 expression confers myeloma stem cell features. Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou,
 RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
RARα2 expression confers myeloma stem cell features Ye Yang, Jumei Shi, Giulia Tolomelli, Hongwei Xu, Jiliang Xia, He Wang, Wen Zhou, Yi Zhou, Satyabrata Das, Zhimin Gu, Dana Levasseur, Fenghuang Zhan,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
doi:10.1038/nature10648 Supplementary Figure 1: Timing of CHIR99021 exposure determines induction of FOXA2/LMX1A midbrain floor plate precursors. a) Immunocytochemical analysis of FOXA2(red)/LMX1A(green)
The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT
 Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Material and Methods RT-PCR Primers The following primers were used for RT-PCR: GAPDH forward (Fw) 5 -GAA GGT GAA GGT CGG AGT C-3, GAPDH reverse (Rev) 5 -GAA GAT GGT GAT GGG ATT TC-3, GPX3
Supplementary Online Content
 Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
Supplementary Online Content Chang K, Taggart MW, Reyes-Uribe L, et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. Published online April 16, 2018. doi:10.1001/jamaoncol.2018.1482
NES: normalized enrichment score (analyzed using KEGG pathway gene sets in the GSEA software); FDR:
 Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Supplementary table1. List of gene sets with simultaneously altered the enrichment upon SAP18 overexpression and knockdown. Gene sets enriched by SAP18 and reduced by shsap18 SAP18 against LacZ shsap18
Product Name Code Size Application
 6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
6 His monoclonal antibody CSB-MA000011M0m 100μg ELISA, WB AADAT Antibody CSB-PA843276LA01HU 100μg ELISA, IHC AARSD1 Antibody CSB-PA863119LA01HU 100μg ELISA, WB, IHC AASDHPPT Antibody CSB-PA865121LA01HU
Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing.
 Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Supplementary Figure 1 Correlation of nonsilent somatic mutations with the age and the disease stage of 25 people with NKTCL, subjected to whole-exome sequencing. (a) Distribution of non-silent somatic
Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal
 Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
Supplemental Information Jmjd1c is Required for MLL-AF9 and HOXA9 Mediated AML Stem Cell Self- Renewal Nan Zhu, Mo Chen, Rowena Eng, Joshua DeJong, Amit U. Sinha, Noushin F. Rahnamay, Richard Koche, Fatima
 SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION MicroRNA-195 attenuates Epithelial-Mesenchymal-Transition (EMT) in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1 Richa Singh, Vikas Yadav, Sachin Kumar and Neeru
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
SUPPLEMENTARY INFORMATION Supplementary Materials and Methods Cell culture conditions My-La, SR-786, HuT 78 and HH were grown in RPMI 1640 medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10%
NCBI ID Symbol 2 A2M A4GALT A4GNT 8086 AAAS 13 AADAC AADACL AAGAB 15 AANAT 16 AARS AARS AASS 18 ABAT 19 ABCA1
 NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
NCBI ID Symbol 2 A2M 53947 A4GALT 51146 A4GNT 8086 AAAS 13 AADAC 344752 AADACL2 79719 AAGAB 15 AANAT 16 AARS 57505 AARS2 10157 AASS 18 ABAT 19 ABCA1 10349 ABCA10 26154 ABCA12 154664 ABCA13 20 ABCA2 21
Supplementary Table S1 Demographic characteristics of HCC patients
 Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Table S1 Demographic characteristics of HCC patients Age (year; mean ± SD) 53.950 ± 11.802 Gender (number; %) Male 79 (79%) Female 21 (21%) AFP (ng/ml; mean ± SD) 23928.571 ± 79577.084 Tumor
Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis
 S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
S1 of S29 Supplementary Material: Phenotype-Genotype Association Analysis of ACTH-Secreting Pituitary Adenoma and Its Molecular Link to Patient Osteoporosis Renzhi Wang, Yakun Yang, Miaomiao Sheng, Dechao
Supplementary Material for. Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility
 Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
Supplementary Material for Supplementary Material for Lee et al., Page 1 Disruption of Ttll5/Stamp gene in male mice causes sperm malformation and infertility Geun-Shik Lee, Yuanzheng He, Edward J. Dougherty,
 Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Supplementary Information Supplementary Materials and Methods Multiple myeloma specimens Primary bone marrow samples from 116 MM patients and 12 patients without tumors were obtained from the 4th Department
Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent
 Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt 1,2, Sourav Bandyopadhyay 1,3 and Frank McCormick 1 Authors
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in. and *Christina Chan
 Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma Aritro Nath 1, Irene Li 2, Lewis R. Roberts 3 1, 2, 4 and *Christina Chan 1 Genetics Program,
SUPPLEMENTARY INFORMATION. Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie
 SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
SUPPLEMENTARY INFORMATION Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8 + T cells Michael Sun, Ngoc Ha, Duc-Hung Pham, Megan Frederick, Bandana Sharma, Chie Naruse, Masahide Asano,
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells
 Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee,
Supplementary Materials for
 www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
www.sciencemag.org/content/350/6266/1383/suppl/dc1 Supplementary Materials for RNA polymerase II associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II Ming Yu, Wenjing
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically Useful
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Lung Cancer Gene qpcr Array For focused group profiling of human lung cancer related genes expression Cat. No. QG071-A (6 x 96-well plate, Format
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma. Supplement Figure 1. Regulation of PGC1α expression in cultured podocytes.
 Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Increasing the level of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 α in podocytes results in collapsing glomerulopathy Supplement Figure 1. Regulation of PGC1α expression in cultured
Analysis Summary: Any Allergy
 Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Analysis Summary: Any Allergy Phenotype Description The any_allergy phenotype includes cases identified from the union of the following sources: Allergies and Asthma survey: An allergic reaction (with
Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein
 1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
1 Supplementary material Blood hsa-mir-122-5p and hsa-mir-885-5p levels associate with fatty liver and related lipoprotein metabolism The Young Finns Study Emma Raitoharju, Ilkka Seppälä, Leo-Pekka Lyytikäinen,
Supplementary Materials for
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Supplementary Materials for PD-1 Marks Dysfunctional Regulatory T cells in Malignant Gliomas Daniel E. Lowther 1,, Brittany A. Goods 2,, Liliana E. Lucca 1,, Benjamin A.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 Supplementary Materials for PD-1 Marks Dysfunctional Regulatory T cells in Malignant Gliomas Daniel E. Lowther 1,, Brittany A. Goods 2,, Liliana E. Lucca 1,, Benjamin A.
Nature Medicine doi: /nm.2457
 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 ature Medicine doi:10.1038/nm.2457 Table S1 - Candidate genes
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we
 SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
SUPPLEMENTARY MATERIALS AND METHODS: Generation of conditional Ada3 knockout targeting construct-to generate a conditional targeting construct, we examined the genomic structure of the mouse Ada3 gene.
500 New CUSABIO Antibodies at MedStore!
 BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
BioMart 500 New CUSABIO Antibodies at MedStore! Purchase two or more units from the attached list of antibodies at MedStore and receive an extra unit for FREE! Valid for purchases made until January 31
GeneCopoeia provides five qpcr array formats (A, B, C, D, and E) suitable for use with the following realtime
 GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
GeneCopoeia TM Expressway to Discovery ExProfile TM Human Breast Cancer Gene qpcr Array For focused group profiling of human breast cancer related genes expression Cat. No. QG063-A (6 x 96-well plate,
Petri Wiklund, Xiaobo Zhang, Satu Pekkala, Reija Autio, Lingjia Kong, Yifan Yang, Sirkka
 Insulin resistance is associated with altered amino acid metabolism and adipose tissue dysfunction in normoglycemic women. Petri Wiklund, Xiaobo Zhang, Satu Pekkala, Reija Autio, Lingjia Kong, Yifan Yang,
Insulin resistance is associated with altered amino acid metabolism and adipose tissue dysfunction in normoglycemic women. Petri Wiklund, Xiaobo Zhang, Satu Pekkala, Reija Autio, Lingjia Kong, Yifan Yang,
Supplementary Figure 1.
 Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Supplementary Figure 1. a 0.772% 0.0126% CD4 2.93% Lineage CD44 CD8 CD25 c-kit b WT Ly5.1 Bone marrow 1:1 Ltbr +/- or Ltbr -/- Ly5.2 10Gy WT % of CD45.2+ ETP among total ETPs 80 60 40 20 0 Ltbr +/- n.s.
Burden of Disease Variants in Participants of the Long Life Family Study. Supplemental Online Material
 Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
Burden of Disease Variants in Participants of the Long Life Family Study Supplemental Online Material Note that in Tables S1-S3 the following scheme was used to denote each Genetic Risk Score (GRS) GRS1:
IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion
 Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Material IL-7-dependent STAT1 activation limits homeostatic CD4 T cell expansion Cecile Le Saout 1, Megan A. Luckey 2, Alejandro V. Villarino 3, Mindy Smith 1, Rebecca B. Hasley 1, Timothy
Supplemental Information. A p53 Super-tumor Suppressor Reveals a Tumor. Suppressive p53-ptpn14-yap Axis. in Pancreatic Cancer
 Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
Cancer Cell, Volume 32 Supplemental Information A p53 Super-tumor Suppressor Reveals a Tumor Suppressive p53-ptpn14-yap Axis in Pancreatic Cancer Stephano S. Mello, Liz J. Valente, Nitin Raj, Jose A. Seoane,
SUPPLEMENTARY INFORMATION. Supplemenary Figures Figure S1
 doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
doi: 10.1038/nature05939 SUPPLEMENTARY INFORMATION Supplemenary Figures Figure S1 wt MEF.1 wt MEF.2 wt MEF.Myc.1 wt MEF.Myc.2 wt MEF.Ras.1 wt MEF.Ras.2 wt MEF.E1A/Ras.1 wt MEF.E1A/Ras.2 p53-/- MEF.E1A/Ras.1
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data. Primer. direction. Forward
 Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplementary Table 1. Primer sequences used for RT-qPCR validation of the microarray data Gene Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) Cell death-inducing DFFAlike effector a (Cidea) Peroxisome
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression.
 Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental Table 1. Oligonucleotides used to identify H-RAS, K-RAS and N-RAS mutations and PTEN gene expression. Gene Exon Forward primer Reverse primer Temp ( C) HRAS 2-3 ATGACGGAATATAAGCTGGT ATGGCAAACACACACAGGAA
Supplemental file 3. All 306 mapped IDs collected by IPA program. Supplemental file 6. The functions and main focused genes in each network.
 LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
LIST OF SUPPLEMENTAL FILES Supplemental file 1. Primer sets used for qrt-pcr. Supplemental file 2. All 1305 differentially expressed genes. Supplemental file 3. All 306 mapped IDs collected by IPA program.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros
SUPPLEMENTARY INFORMATION doi:10.1038/nature20792 Sanger Mouse Genetics Project participants Agnieszka Swiatkowska, Amy Gates, Angela Green, Anna Karin Gerdin, Brendan Doe, Carl Shannon, Carmen Ballesteros
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1
 Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
Τα ηπατικά επίπεδα του FOXP3 mrna στη χρόνια ηπατίτιδα Β εξαρτώνται από την έκφραση των οδών Fas/FasL και PD-1/PD-L1 Γ. Γερμανίδης, Ν. Αργέντου, Θ. Βασιλειάδης, Κ. Πατσιαούρα, Κ. Μαντζούκης, A. Θεοχαρίδου,
In utero vitamin D deficiency increases airway smooth muscle mass and impairs lung
 1 Supplemental Information 2 Title 3 4 In utero vitamin D deficiency increases airway smooth muscle mass and impairs lung function 5 Authors 6 7 1 Rachel E Foong, 1 Anthony Bosco, 1 Anya C Jones, 1 Alex
1 Supplemental Information 2 Title 3 4 In utero vitamin D deficiency increases airway smooth muscle mass and impairs lung function 5 Authors 6 7 1 Rachel E Foong, 1 Anthony Bosco, 1 Anya C Jones, 1 Alex
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
doi:10.1038/nature11847 WWW.NATURE.COM/NATURE 1 WWW.NATURE.COM/NATURE 2 WWW.NATURE.COM/NATURE 3 WWW.NATURE.COM/NATURE 4 WWW.NATURE.COM/NATURE 5 WWW.NATURE.COM/NATURE 6 WWW.NATURE.COM/NATURE 7 a SSC singlets
GeneTex Sonderaktion: Custom Antibody Panels
 (Stand 01/2016) GeneTex Sonderaktion: Custom Antibody Panels Fünf Antikörper (in der 25 µl-größe) zum Sonderpreis von nur 420,00 (statt 545,00 ). Bitte geben Sie bei Ihrer Bestellung die Angebotsnummer
(Stand 01/2016) GeneTex Sonderaktion: Custom Antibody Panels Fünf Antikörper (in der 25 µl-größe) zum Sonderpreis von nur 420,00 (statt 545,00 ). Bitte geben Sie bei Ihrer Bestellung die Angebotsnummer
Supplementary Information for. RRM2B Suppresses Activation of the Oxidative Stress Pathway and is. Up-regulated by P53 During Senescence
 Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
Supplementary Information for RRM2B Suppresses Activation of the Oxidative Stress Pathway and is Up-regulated by P53 During Senescence Mei-Ling Kuo 1,**, Alexander J. Sy 1, Lijun Xue 1, Martin Chi 1, Michelle
(A) Rounds of replating (R) of MLL-AF9 transformed LSK colonies from initiation
 upplemental Figure. (A) Rounds of replating (R) of LL-AF transformed LK colonies from initiation experiments (.E. of triplicate experiments, p=.)., p
upplemental Figure. (A) Rounds of replating (R) of LL-AF transformed LK colonies from initiation experiments (.E. of triplicate experiments, p=.)., p
Supplemental Table 1. ICD-9-CM codes and ATC codes used in this study
 Supplemental Table 1. ICD-9-CM codes and ATC codes used in this study Comorbidities ICD-9-CM codes Medication ATC codes Diabetes 250 Angiotensin-converting enzyme inhibitors C09AA Ischemic heart disease
Supplemental Table 1. ICD-9-CM codes and ATC codes used in this study Comorbidities ICD-9-CM codes Medication ATC codes Diabetes 250 Angiotensin-converting enzyme inhibitors C09AA Ischemic heart disease
Christopher Stephen Inchley, 2015
 Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Christopher Stephen Inchley, 2015 Series of dissertations submitted to the Faculty of Medicine, University of Oslo No. 2009 ISBN 978-82-8264-990-2 All rights reserved. No part of this publication may be
Η γενετική πολυπλοκότητα του καρκίνου του παγκρέατος
 Η γενετική πολυπλοκότητα του καρκίνου του παγκρέατος Δρ Αθανάσιος Αθανασιάδης Αθανάσιος Αθανασιάδης Δεν έχω αντικρουόμενα συμφέροντα προς γνωστοποίηση Καρκίνος παγκρέατος Θνητότης Επίπτωση Καθυστερημένη
Η γενετική πολυπλοκότητα του καρκίνου του παγκρέατος Δρ Αθανάσιος Αθανασιάδης Αθανάσιος Αθανασιάδης Δεν έχω αντικρουόμενα συμφέροντα προς γνωστοποίηση Καρκίνος παγκρέατος Θνητότης Επίπτωση Καθυστερημένη
CD4 + T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2
 Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland
Supplementary Information, Boucheron et al., 1 Supplementary Information for the manuscript T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2 Nicole Boucheron 1,11, Roland
Supplementary Table 1. Composition of the diets
 Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Supplementary Table 1. Composition of the diets Standard diet High Fat diet Energy content (kcal/kg) 2508 3701 Protein (g/kg) 200 170 Carbohydrate (g/kg) 720 670 Total fat (g/kg) 80 160 Saturated fatty
Original Article Transcriptome analysis of primary aldosteronism in adrenal glands and controls
 Int J Clin Exp Pathol 2017;10(9):10009-10018 www.ijcep.com /ISSN:1936-2625/IJCEP0059197 Original Article Transcriptome analysis of primary aldosteronism in adrenal glands and controls Chenlong Chu 1*,
Int J Clin Exp Pathol 2017;10(9):10009-10018 www.ijcep.com /ISSN:1936-2625/IJCEP0059197 Original Article Transcriptome analysis of primary aldosteronism in adrenal glands and controls Chenlong Chu 1*,
Santa Cruz Biotechnology, Inc.
 sc-24 X sc-40 X sc-41 X sc-42 X sc-44 X sc-45 X sc-46 X sc-48 X sc-50 X sc-52 X sc-59 X sc-61 X sc-70 X sc-71 X sc-73 X sc-74 X sc-109 X sc-110 X sc-111 X sc-112 X sc-113 X sc-114 X sc-122 X sc-126 X sc-131
sc-24 X sc-40 X sc-41 X sc-42 X sc-44 X sc-45 X sc-46 X sc-48 X sc-50 X sc-52 X sc-59 X sc-61 X sc-70 X sc-71 X sc-73 X sc-74 X sc-109 X sc-110 X sc-111 X sc-112 X sc-113 X sc-114 X sc-122 X sc-126 X sc-131
SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in
 1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary
Long intergenic non-coding RNA expression signature in human breast cancer
 Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
Long intergenic non-coding RNA expression signature in human breast cancer Yanfeng Zhang 1,5,#, Erin K. Wagner 2,#, Xingyi Guo 1, Isaac May 3, Qiuyin Cai 1, Wei Zheng 1, Chunyan He 2,4,*, Jirong Long 1,*
encouraged to use the Version of Record that, when published, will replace this version. The most /BCJ
 Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Biochemical Journal: this is an Accepted Manuscript, not the final Version of Record. You are encouraged to use the Version of Record that, when published, will replace this version. The most up-to-date
Supplementary Materials for
 www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
www.sciencemag.org/content/351/6273/613/suppl/dc1 Supplementary Materials for Foxc1 reinforces quiescence in self-renewing hair follicle stem cells Li Wang, Julie A. Siegenthaler, Robin D. Dowell, Rui
Zhang et al., Supplemental figures, Methods, References and Tables
 1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
1 2 Zhang et al., Supplemental figures, Methods, References and Tables Supplemental figures 3 4 5 6 7 8 9 10 11 14 15 16 17 18 19 20 Supplemental Figure 1. Geminin interacts with FoxO3 (A and B) Cell lysates
ΚΛΑΙΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ ΜΟΡΙΑΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ ΜΟΡΙΑΚΗ ΓΕΝΕΣΙΚΗ ΜΟΡΙΑΚΗ ΜΙΚΡΟΒΙΟΛΟΓΙΑ. Χρόνοσ Απάντθςθσ
 ΚΛΑΙΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ Χρωμοςωμικι ανάλυςθ αμνιακοφ υγροφ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ CVS (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ εμβρυϊκοφ αίματοσ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ
ΚΛΑΙΚΗ ΚΤΣΣΑΡΟΓΕΝΕΣΙΚΗ Χρωμοςωμικι ανάλυςθ αμνιακοφ υγροφ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ CVS (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ εμβρυϊκοφ αίματοσ (καρυότυποσ και PCR) Χρωμοςωμικι ανάλυςθ
Ι Α ΚΑΛΙΑ Θ ΩΡΗΣΙΚΩΝ ΜΑΘΗΜΑΣΩΝ ΠΑΘΟΛΟΓΙΑ ΙΙ
 Ι Α ΚΑΛΙΑ Θ ΩΡΗΣΙΚΩΝ ΜΑΘΗΜΑΣΩΝ ΠΑΘΟΛΟΓΙΑ ΙΙ 1 ΑΘΗ Ε Η Α Α Χ ΗΦ ΩΝ Η 2 Η Η Η Η Η K Ω Η SGOT-AST SGPT-ALT ( ω ό ) Χ Η Χ Η ALP GT Χ Θ Η Η Θ Η Η ALT (SGPT) and AST (SGOT) Enzymes found in Hepatocytes Released
Ι Α ΚΑΛΙΑ Θ ΩΡΗΣΙΚΩΝ ΜΑΘΗΜΑΣΩΝ ΠΑΘΟΛΟΓΙΑ ΙΙ 1 ΑΘΗ Ε Η Α Α Χ ΗΦ ΩΝ Η 2 Η Η Η Η Η K Ω Η SGOT-AST SGPT-ALT ( ω ό ) Χ Η Χ Η ALP GT Χ Θ Η Η Θ Η Η ALT (SGPT) and AST (SGOT) Enzymes found in Hepatocytes Released
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
www.sciencesignaling.org/cgi/content/full/9/444/ra87/dc1 Supplementary Materials for MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells Atilgan Yilmaz, Chandramohan
Analysis Summary: Hypermobility Beighton Score
 Analysis Summary: Hypermobility Beighton Score Phenotype Description The beighton_hypermobility_qtl phenotype comes from: morphology survey: Can you bring your right thumb to reach your forearm (see above
Analysis Summary: Hypermobility Beighton Score Phenotype Description The beighton_hypermobility_qtl phenotype comes from: morphology survey: Can you bring your right thumb to reach your forearm (see above
Differentially expressed genes of HEK293/HeLa Myst4 sirna vs. controls.
 Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Supplemental data - Content Supplemental figures Figure 1 Figure 2 Figure 3 Figure 4 Western blot analysis of MYST4. Expression pattern of MYST4 isoforms. Differentially expressed genes of HEK293/HeLa
Estimation of grain boundary segregation enthalpy and its role in stable nanocrystalline alloy design
 Supplemental Material for Estimation of grain boundary segregation enthalpy and its role in stable nanocrystalline alloy design By H. A. Murdoch and C.A. Schuh Miedema model RKM model ΔH mix ΔH seg ΔH
Supplemental Material for Estimation of grain boundary segregation enthalpy and its role in stable nanocrystalline alloy design By H. A. Murdoch and C.A. Schuh Miedema model RKM model ΔH mix ΔH seg ΔH
ΔΙΑΤΡΙΒΗ. για την απόκτηση ΜΕΤΑΠΤΥΧΙΑΚΟΥ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ
 ΤΜΗΜΑ ΦΑΡΜΑΚΕΥΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ ΦΑΡΜΑΚΕΥΤΙΚΕΣ ΕΠΙΣΤΗΜΕΣ ΚΑΙ ΤΗ ΤΕΧΝΟΛΟΓΙΑ ΔΙΑΤΡΙΒΗ για την απόκτηση ΜΕΤΑΠΤΥΧΙΑΚΟΥ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ Φαρμακογονιδιωματική και λειτουργική μελέτη
ΤΜΗΜΑ ΦΑΡΜΑΚΕΥΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ ΣΤΙΣ ΦΑΡΜΑΚΕΥΤΙΚΕΣ ΕΠΙΣΤΗΜΕΣ ΚΑΙ ΤΗ ΤΕΧΝΟΛΟΓΙΑ ΔΙΑΤΡΙΒΗ για την απόκτηση ΜΕΤΑΠΤΥΧΙΑΚΟΥ ΔΙΠΛΩΜΑΤΟΣ ΕΙΔΙΚΕΥΣΗΣ Φαρμακογονιδιωματική και λειτουργική μελέτη
Το mir-29a ως βιοδείκτης στην υπερτροφική μυοκαρδιοπάθεια. Δ. Ντέλιος 1,2
 Το mir-29a ως βιοδείκτης στην υπερτροφική μυοκαρδιοπάθεια Δ. Ντέλιος 1,2 Υπερτροφική μυοκαρδιοπάθεια - 1:500 στο γενικό πληθυσμό Πρωτεΐνες του σαρκομεριδίου ΜΥΗ7, MYBPC3, TNNI3, TNNT2, TPM1, MYL3, MYL2
Το mir-29a ως βιοδείκτης στην υπερτροφική μυοκαρδιοπάθεια Δ. Ντέλιος 1,2 Υπερτροφική μυοκαρδιοπάθεια - 1:500 στο γενικό πληθυσμό Πρωτεΐνες του σαρκομεριδίου ΜΥΗ7, MYBPC3, TNNI3, TNNT2, TPM1, MYL3, MYL2
Supplementary Information
 Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary Information Recurrent PAX3-MAML3 Fusion in Biphenotypic Sinonasal Sarcoma Xiaoke Wang 1,, Krista L. Bledsoe 2,, Rondell P. Graham 1,, Yan W. Asmann 3, David S. Viswanatha 1, Jean E. Lewis
Supplementary data to: The mir-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2
 Supplementary data to: The mir-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB and ZEB2 Sun-Mi Park, Arti B. Gaur 3, Ernst Lengyel 2 and Marcus
Supplementary data to: The mir-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB and ZEB2 Sun-Mi Park, Arti B. Gaur 3, Ernst Lengyel 2 and Marcus
ΖΕΡΔΑΛΗΣ ΣΩΤΗΡΙΟΣ ΤΟ ΟΥΤΙ ΣΤΗ ΒΕΡΟΙΑ (1922-ΣΗΜΕΡΑ) ΘΕΣΣΑΛΟΝΙΚΗ 2005 1
 (1922- ) 2005 1 2 .1.2 1.1.2-3 1.2.3-4 1.3.4-5 1.4.5-6 1.5.6-10.11 2.1 2.2 2.3 2.4.11-12.12-13.13.14 2.5 (CD).15-20.21.22 3 4 20.,,.,,.,.,,.,.. 1922., (= )., (25/10/2004), (16/5/2005), (26/1/2005) (7/2/2005),,,,.,..
(1922- ) 2005 1 2 .1.2 1.1.2-3 1.2.3-4 1.3.4-5 1.4.5-6 1.5.6-10.11 2.1 2.2 2.3 2.4.11-12.12-13.13.14 2.5 (CD).15-20.21.22 3 4 20.,,.,,.,.,,.,.. 1922., (= )., (25/10/2004), (16/5/2005), (26/1/2005) (7/2/2005),,,,.,..
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ
 ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΚΥΤΤΑΡΟΓΕΝΕΤΙΚΗΣ ΚΑΙ ΜΟΡΙΑΚΗΣ ΓΕΝΕΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΓΕΝΕΤΙΚΗ ΤΟΥ ΑΝΘΡΩΠΟΥ» ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ «Σύγκριση μελετών
ΠΑΝΕΠΙΣΤΗΜΙΟ ΘΕΣΣΑΛΙΑΣ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΥΓΕΙΑΣ ΤΜΗΜΑ ΙΑΤΡΙΚΗΣ ΕΡΓΑΣΤΗΡΙΟ ΚΥΤΤΑΡΟΓΕΝΕΤΙΚΗΣ ΚΑΙ ΜΟΡΙΑΚΗΣ ΓΕΝΕΤΙΚΗΣ ΠΡΟΓΡΑΜΜΑ ΜΕΤΑΠΤΥΧΙΑΚΩΝ ΣΠΟΥΔΩΝ «ΓΕΝΕΤΙΚΗ ΤΟΥ ΑΝΘΡΩΠΟΥ» ΔΙΠΛΩΜΑΤΙΚΗ ΕΡΓΑΣΙΑ «Σύγκριση μελετών
Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia
 SUPPLEMENTAL MATERIAL FOR: Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia Matthew T. Witkowski, Luisa Cimmino, Yifang Hu, Thomas Trimarchi,
SUPPLEMENTAL MATERIAL FOR: Activated Notch counteracts Ikaros tumor suppression in mouse and human T cell acute lymphoblastic leukemia Matthew T. Witkowski, Luisa Cimmino, Yifang Hu, Thomas Trimarchi,
Cellular Physiology and Biochemistry
 Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Original Paper 2016 The Author(s). 2016 Published The Author(s) by S. Karger AG, Basel Published online: November 25, 2016 www.karger.com/cpb Published by S. Karger AG, Basel 486 www.karger.com/cpb Accepted:
Study of the gene expression profile in the bone tissue with kidney yin deficiency syndromes in primary oste oporosis
 203 2 9 2 Chin J OsteoporosDecember 203 Vol 9No. 2 Published online www. wanfangdate. com. cn doi0. 3969 /j. issn. 006-708. 203. 2. 00 25 2 2 *. 350003 2. 350007 R68 A 006-7082032-25-05 3 3 3 3 4 2378
203 2 9 2 Chin J OsteoporosDecember 203 Vol 9No. 2 Published online www. wanfangdate. com. cn doi0. 3969 /j. issn. 006-708. 203. 2. 00 25 2 2 *. 350003 2. 350007 R68 A 006-7082032-25-05 3 3 3 3 4 2378
Supplemental Materials and Results
 Supplementary Figures and Legends Supplemental Materials and Results Supplementary Figure 1. Metabolic parameters in Ad-mKO mice A. TG content in skeletal muscle of Ctrl and Ad-mKO mice on chow diet (CD)
Supplementary Figures and Legends Supplemental Materials and Results Supplementary Figure 1. Metabolic parameters in Ad-mKO mice A. TG content in skeletal muscle of Ctrl and Ad-mKO mice on chow diet (CD)
Table S1A. Generic EMT signature for tumour
 Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
Table S1A. Generic EMT signature for tumour Index Gene Symbol Gene Title GO Molecular Function Weight Weight.Zt ransform ed Category In Generic Cell Line EMT Signature = 1 1 KRT19 Keratin, type I cytoskeletal
Systematic Analysis of Candidate Genes for Alzheimer s Disease in a French, Genome-Wide Association Study
 Journal of Alzheimer s Disease 20 (2010) 1 13 1 IOS Press Supplementary Material Systematic Analysis of Candidate Genes for Alzheimer s Disease in a French, Genome-Wide Association Study Geoffroy Laumet
Journal of Alzheimer s Disease 20 (2010) 1 13 1 IOS Press Supplementary Material Systematic Analysis of Candidate Genes for Alzheimer s Disease in a French, Genome-Wide Association Study Geoffroy Laumet
http / / cjbmb. bjmu. edu. cn Chinese Journal of Biochemistry and Molecular Biology RT-PCR Runx d PCR RT-qPCR Western BMPs Wnt Ihh
 ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 6 27 6 540 ~ 547 micrornas * / / 250062 microrna mrna β- RT-PCR Runx2 0 7 14 d micrornas
ISSN 1007-7626 CN 11-3870 / Q http / / cjbmb bjmu edu cn Chinese Journal of Biochemistry and Molecular Biology 2011 6 27 6 540 ~ 547 micrornas * / / 250062 microrna mrna β- RT-PCR Runx2 0 7 14 d micrornas
Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development
 Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development Isabelle Buisson, Ronan Le Bouffant, Mélinée Futel, Jean-François Riou, Muriel Umbhauer To cite this version:
Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development Isabelle Buisson, Ronan Le Bouffant, Mélinée Futel, Jean-François Riou, Muriel Umbhauer To cite this version:
5- CACGAAACTACCTTCAACTCC-3 beta actin-r 5- CATACTCCTGCTTGCTGATC-3 GAPDH-F GAPDH-R
 Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Table S1 Primers used in this work LncPPARδ-F 5- AGGATCCCAAGGCAGAAGTT -3 LncPPARδ-R 5- GAATAGAAAGGCGGGAAAGG -3 PPARδ-F 5- CACCCAGTGTCCTTCCATCT -3 PPARδ-R 5- CAGAATTGGGTGTGCTGCTT -3 ADRP-F 5- ACAACCGAGTGTGGTGACTC
Metabolite. Metabolite conc.: μm. Blood. Blood. mg/l
 55-methanol (CAS# 67-56-1) Table 1a. Sub-lethal acute poisoning (single dose): Clinical observations (time related) Ref Reference (linked to full source) age/sex category Dose: g Notes (case, dose, time)
55-methanol (CAS# 67-56-1) Table 1a. Sub-lethal acute poisoning (single dose): Clinical observations (time related) Ref Reference (linked to full source) age/sex category Dose: g Notes (case, dose, time)
Expression status of diabetes-associated genes in middle and aged cynomolgus monkeys
 2011 Jun. 32(3): 300 306 CN 53-1040/Q ISSN 0254-5853 Zoological Research DOI 10.3724/SP.J.1141.2011.03300 1, 1, 1, 2, 1, 1, 1, 2, 1,2,* (1. 510260 2. 510555) 10 138, (FPG) (TCHO) (TG) (HDL-C) (LDL-C) ;
2011 Jun. 32(3): 300 306 CN 53-1040/Q ISSN 0254-5853 Zoological Research DOI 10.3724/SP.J.1141.2011.03300 1, 1, 1, 2, 1, 1, 1, 2, 1,2,* (1. 510260 2. 510555) 10 138, (FPG) (TCHO) (TG) (HDL-C) (LDL-C) ;
 Νόσος Parkinson-Νέοι ορίζοντες Ξηροµερήσιου Γεωργία Νευρολόγος-Μέλος ερευνητικής οµάδας Νευρογενετικής Αίτια της νόσου Parkinson Περιβαλλοντικοί παράγοντες Γενετικοί παράγοντες Γενετική βλάβη (παθογόνα
Νόσος Parkinson-Νέοι ορίζοντες Ξηροµερήσιου Γεωργία Νευρολόγος-Μέλος ερευνητικής οµάδας Νευρογενετικής Αίτια της νόσου Parkinson Περιβαλλοντικοί παράγοντες Γενετικοί παράγοντες Γενετική βλάβη (παθογόνα
In the United States, urothelial carcinoma (UC) of the bladder
 Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology Jeffrey S. Damrauer a,b, Katherine A. Hoadley a,b, David D. Chism a,c, Cheng Fan a,b, Christopher J. Tiganelli
Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology Jeffrey S. Damrauer a,b, Katherine A. Hoadley a,b, David D. Chism a,c, Cheng Fan a,b, Christopher J. Tiganelli
Supplementary information. Somatic SETBP1 mutations in myeloid malignancies
 Supplementary information Somatic SETBP1 mutations in myeloid malignancies Hideki Makishima, Kenichi Yoshida, Nhu Nguyen, Bartlomiej Przychodzen, Masashi Sanada, Yusuke Okuno, Kwok Peng Ng, Kristbjorn
Supplementary information Somatic SETBP1 mutations in myeloid malignancies Hideki Makishima, Kenichi Yoshida, Nhu Nguyen, Bartlomiej Przychodzen, Masashi Sanada, Yusuke Okuno, Kwok Peng Ng, Kristbjorn

